Introduction
This vignette takes the user through some basic scenarios for
exploring, assembling, and validating data sets using
mrgda, introducing you to its standard workflow and
functionality. For more information how to effectively integrate
mrgda into your workflow, visit the
MeRGE Expo.
Source data
We begin by pointing to a source directory containing the source data
(typically STDM or ADaM datasets). read_src_dir will read
in every data file and return a named list of the data objects, as well
as some additional metadata.
src_path <- system.file("example-sdtm", package = "mrgda")
# Read in all XPT files in source directory
src_list <- mrgda::read_src_dir(src_path, .file_types = "xpt")Exploring the source data
While it is common practice to use View() for inspecting
dataframes or matrix-like R objects, mrgda
provides a convenient tool for exploring your entire source
directory.
mrgda::v(src_list)v() facilitates manual inspections, such as confirming
data integrity for a specific subject or over specified date range. Note
that v() runs a shiny app in the Rstudio
viewer pane as background process, allowing you to retain control over
your R console:
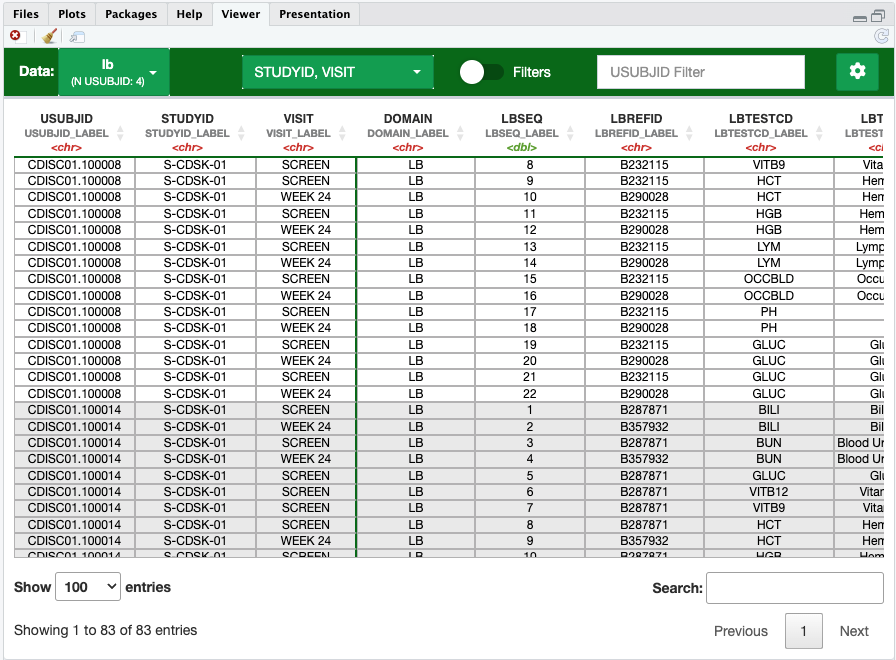
Some features include:
- Allows you to seamlessly transition between any of the datasets in your list (you can also pass in a single dataframe)
- Apply global subject filters, filtering each of the datasets in
src_listto only contain the specified subject ID - Apply column filters per dataset
- Groups the data by the specified or detected
.subject_col, helping to differentiate between subject records. - Column Organization
- “Freeze” or lock columns in place while scrolling horizontally
- Relocate columns to another position
- Other formatting options related to readability and presentation of column label attributes
Query source list
In addition to the convenient viewing and filtering methods provided
by v(), you can also query the entire source directory for
a specific character string via query_src_list():
# Summarize source directory
query_src_list(src_list, .string = "RACE")
#> # A tibble: 2 × 3
#> DOMAIN COLUMNS MATCHING
#> <chr> <chr> <chr>
#> 1 lb LBORRES,LBSTRESC,LBSTNRC RACE
#> 2 mrgda_labels COLUMN_NAME,COLUMN_LABEL RACESave assembled data
After you finish assembling your dataset, you can export the compiled
data using write_derived, ensuring both convenience and
reproducibility. This function produces a CSV, an xpt file,
and a dedicated folder housing valuable metadata. This dual export
capability enhances data portability, accommodating various use cases.
The specification file, nm_spec, is meant to contain
information relating to units, abbreviations, decoding options, and more
(see the
MeRGE Expo for more information on this).
nm_spec <- yspec::ys_load(system.file("derived", "pk.yml", package = "mrgda"))
mrgda::write_derived(
.data = derived$nm,
.spec = nm_spec,
.file = rprojroot::find_rstudio_root_file("data", "derived", "pk.csv")
)Other functions
mrgda exports other helpful functions for data creation,
management, and validation. See the
Reference Log for more details.