library(mrgmisc) # for binning and other functions
library(ggplot2) #plotting
library(purrr) # dataset to handle lists
suppressMessages(suppressWarnings(library(dplyr)))Messy plot can’t see individuals well
ggplot(dat, aes(x = Time, y = Conc, group= ID)) +
geom_line() +
facet_wrap(~ID, scales= "free")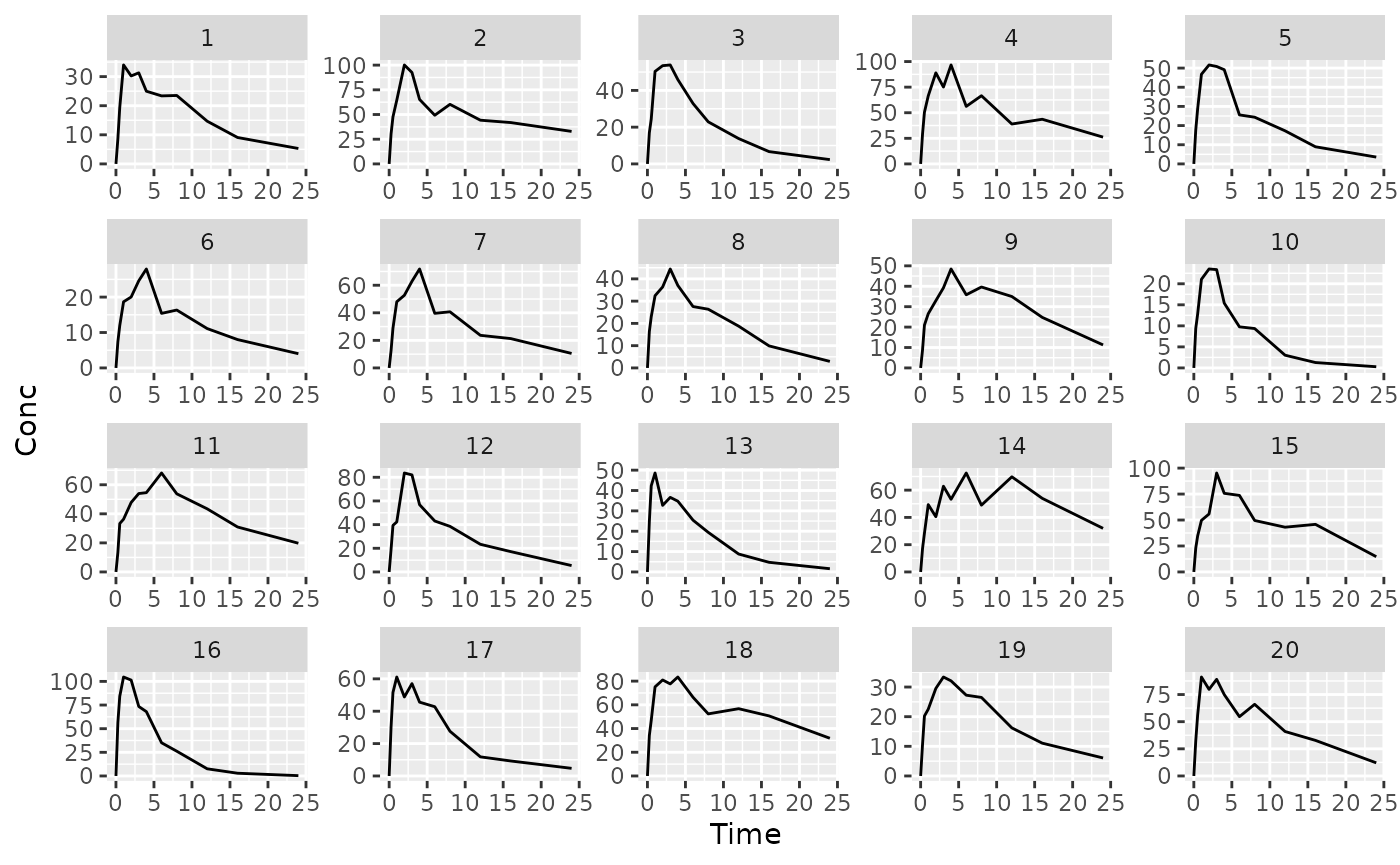
What we’d like to be able to do is split up into ‘bins’ of specified numbers of individuals to then plot out multiple plots
# this will automatically create a column of bins such that the
# specified number of individuals is in each bin, in this case
# 9 ids per bin
# split the original dataset into subdatasets corresponding to each bin (list of dataframes)
split_dat <- dat %>%
mutate(PLOTS = ids_per_plot(ID, 4)) %>% # default is 9 per subplot
split(.[["PLOTS"]])To handle plotting each subdataframe, you need to wrap your normal
ggplot into a function. You will then apply this function with
map to each subdataframe. So in this case it is just like a
normal plot, but wrapped up in a function, which takes 1 argument (the
dataframe) and outputs the plot
p_conc_time <- function(df) {
ggplot(df, aes(x = Time, y = Conc, group= ID)) +
geom_line() +
facet_wrap(~ID, scales= "free")
}To apply the above function we use map from
purrr
## $`1`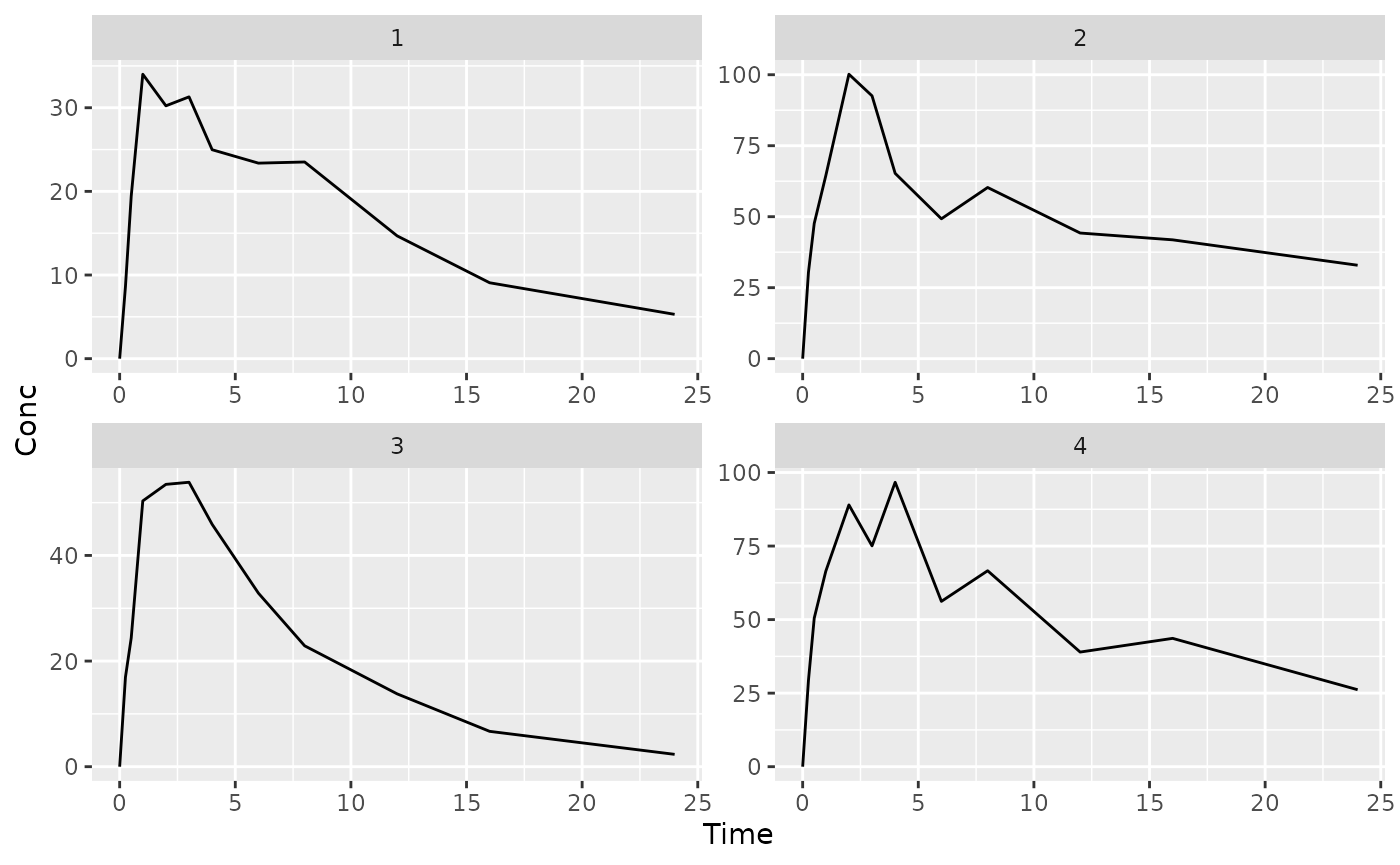
##
## $`2`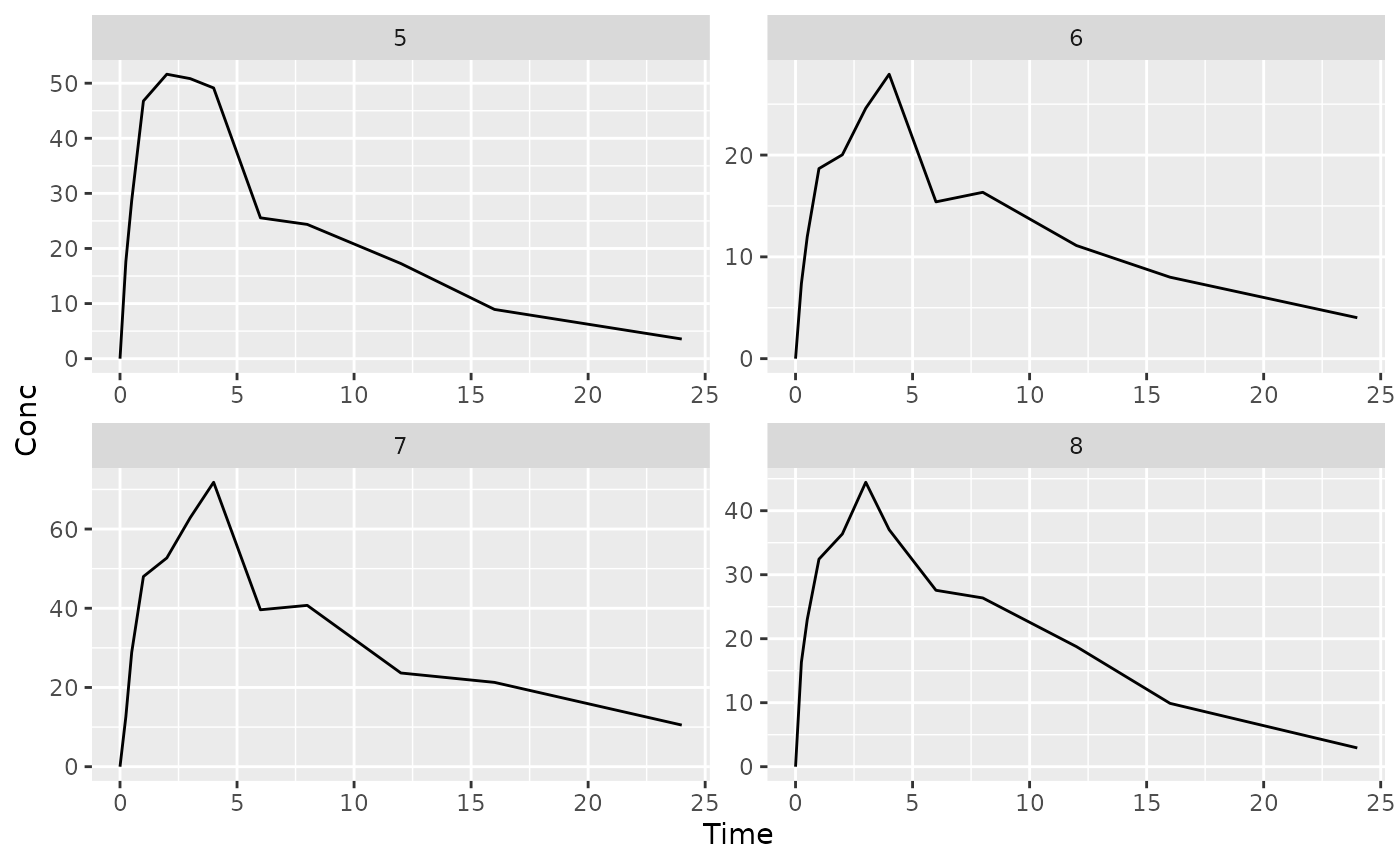
##
## $`3`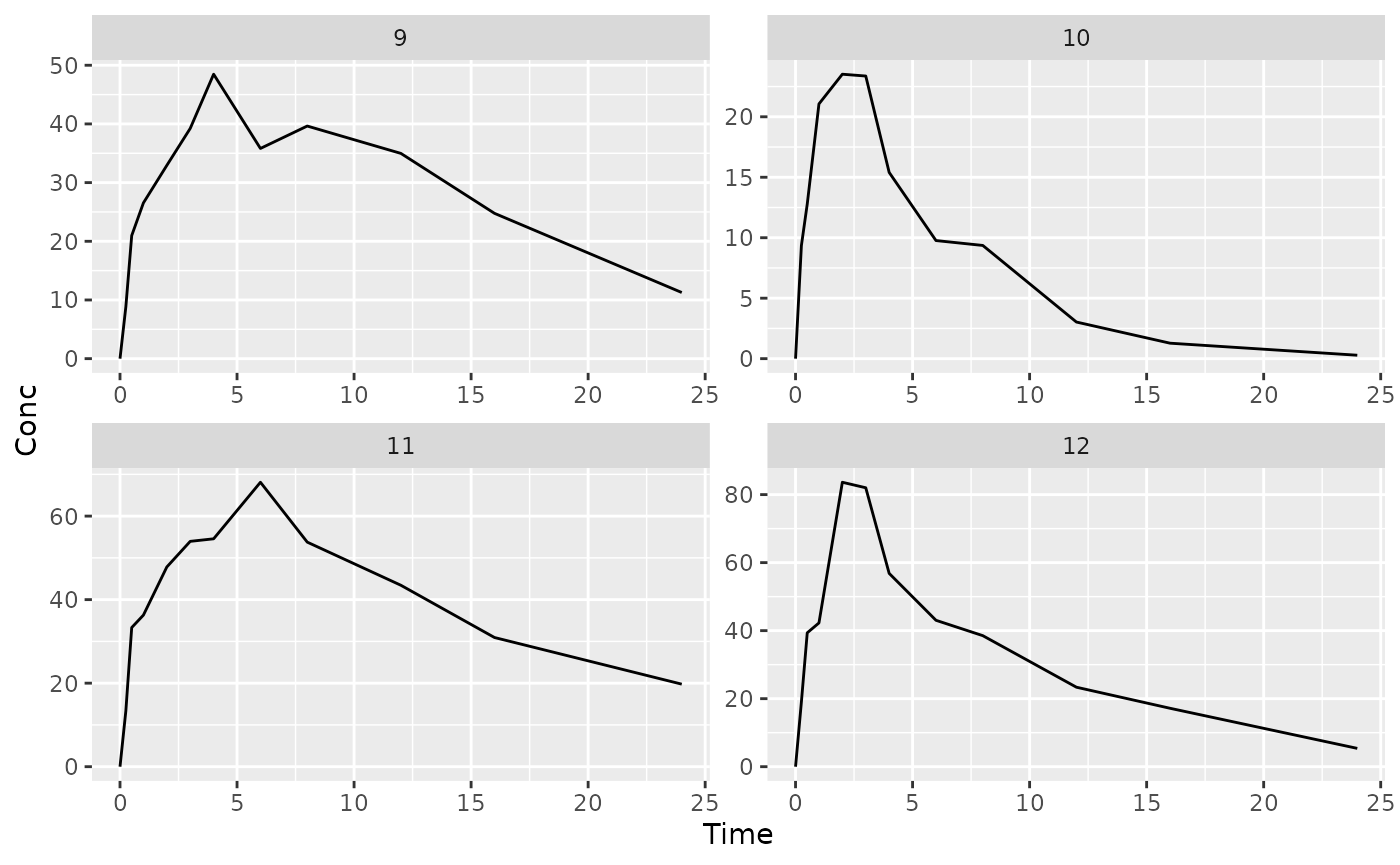
##
## $`4`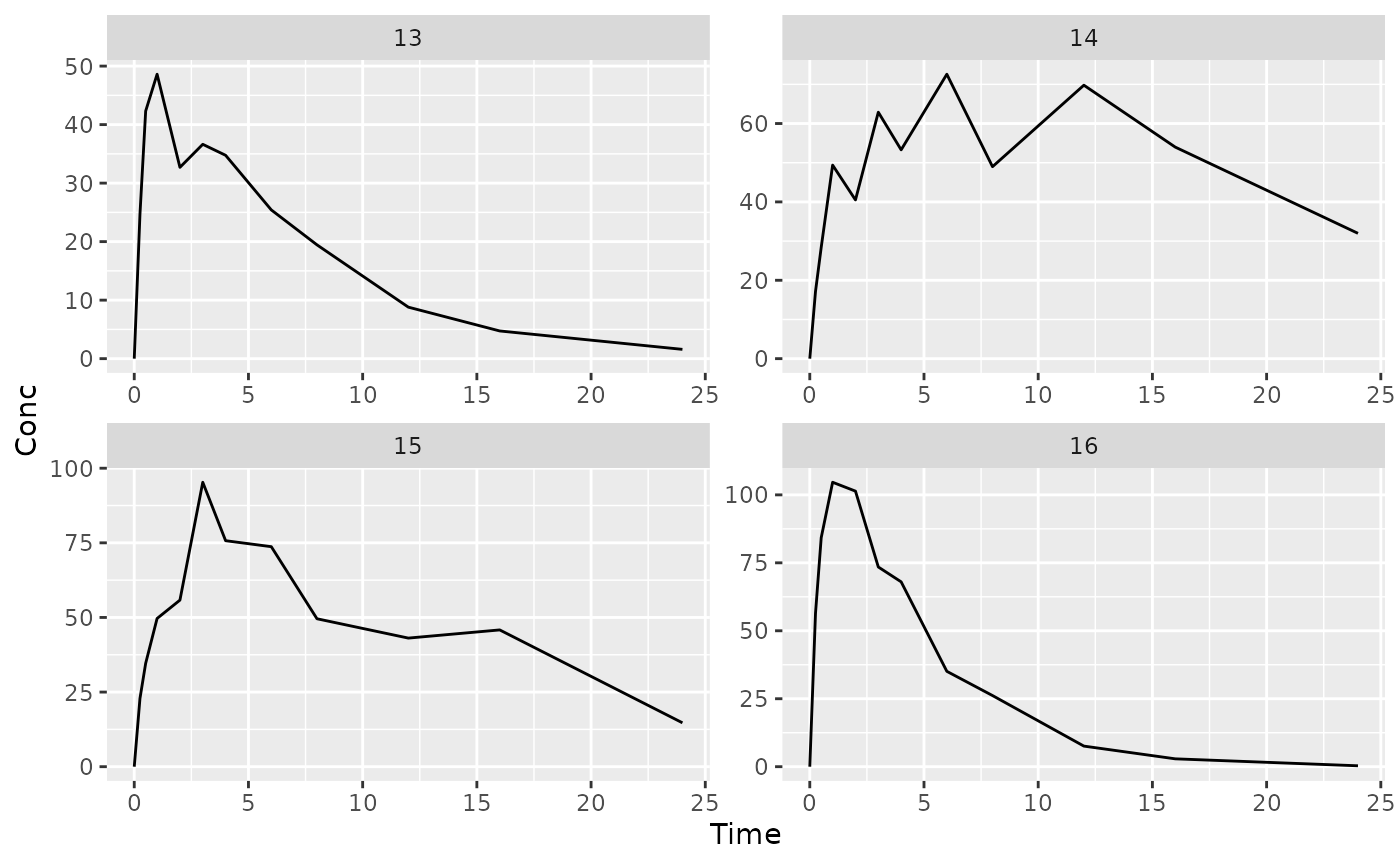
##
## $`5`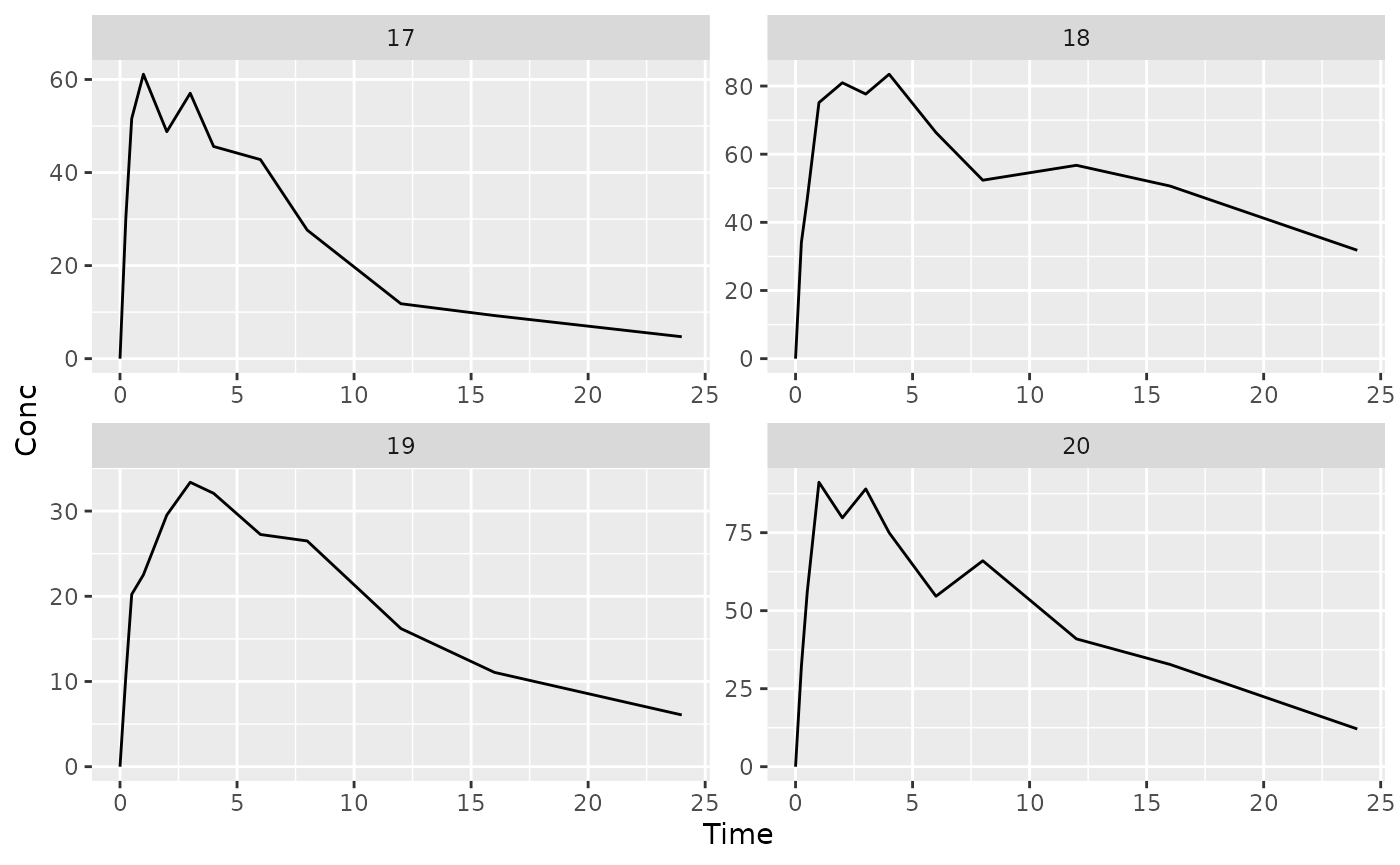
sessioninfo::session_info()## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.4.2 (2024-10-31)
## os Ubuntu 20.04.6 LTS
## system x86_64, linux-gnu
## ui X11
## language en
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz America/New_York
## date 2025-07-30
## pandoc 3.3 @ /usr/bin/ (via rmarkdown)
## quarto 1.3.450 @ /data/home/kylem/bin/quarto
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## bslib 0.9.0 2025-01-30 [2] CRAN (R 4.4.2)
## cachem 1.1.0 2024-05-16 [2] CRAN (R 4.4.2)
## cli 3.6.5 2025-04-23 [2] CRAN (R 4.4.2)
## desc 1.4.3 2023-12-10 [2] CRAN (R 4.4.2)
## digest 0.6.37 2024-08-19 [2] CRAN (R 4.4.2)
## dplyr * 1.1.4 2023-11-17 [2] CRAN (R 4.4.2)
## evaluate 1.0.4 2025-06-18 [2] CRAN (R 4.4.2)
## farver 2.1.2 2024-05-13 [2] CRAN (R 4.4.2)
## fastmap 1.2.0 2024-05-15 [2] CRAN (R 4.4.2)
## fs 1.6.6 2025-04-12 [2] CRAN (R 4.4.2)
## generics 0.1.4 2025-05-09 [2] CRAN (R 4.4.2)
## ggplot2 * 3.5.2 2025-04-09 [2] CRAN (R 4.4.2)
## glue 1.8.0 2024-09-30 [2] CRAN (R 4.4.2)
## gtable 0.3.6 2024-10-25 [2] CRAN (R 4.4.2)
## htmltools 0.5.8.1 2024-04-04 [2] CRAN (R 4.4.2)
## htmlwidgets 1.6.4 2023-12-06 [2] CRAN (R 4.4.2)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.4.2)
## jsonlite 2.0.0 2025-03-27 [2] CRAN (R 4.4.2)
## knitr 1.50 2025-03-16 [2] CRAN (R 4.4.2)
## labeling 0.4.3 2023-08-29 [2] CRAN (R 4.4.2)
## lifecycle 1.0.4 2023-11-07 [2] CRAN (R 4.4.2)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.4.2)
## mrgmisc * 0.3.0 2025-07-30 [1] local
## pillar 1.11.0 2025-07-04 [2] CRAN (R 4.4.2)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.4.2)
## pkgdown 2.1.3 2025-05-25 [2] CRAN (R 4.4.2)
## purrr * 1.1.0 2025-07-10 [2] CRAN (R 4.4.2)
## R6 2.6.1 2025-02-15 [2] CRAN (R 4.4.2)
## ragg 1.4.0 2025-04-10 [2] CRAN (R 4.4.2)
## RColorBrewer 1.1-3 2022-04-03 [2] CRAN (R 4.4.2)
## Rcpp 1.1.0 2025-07-02 [2] CRAN (R 4.4.2)
## rlang 1.1.6 2025-04-11 [2] CRAN (R 4.4.2)
## rmarkdown 2.29 2024-11-04 [2] CRAN (R 4.4.2)
## sass 0.4.10 2025-04-11 [2] CRAN (R 4.4.2)
## scales 1.4.0 2025-04-24 [2] CRAN (R 4.4.2)
## sessioninfo 1.2.3 2025-02-05 [2] CRAN (R 4.4.2)
## systemfonts 1.2.3 2025-04-30 [2] CRAN (R 4.4.2)
## textshaping 1.0.1 2025-05-01 [2] CRAN (R 4.4.2)
## tibble 3.3.0 2025-06-08 [2] CRAN (R 4.4.2)
## tidyselect 1.2.1 2024-03-11 [2] CRAN (R 4.4.2)
## vctrs 0.6.5 2023-12-01 [2] CRAN (R 4.4.2)
## withr 3.0.2 2024-10-28 [2] CRAN (R 4.4.2)
## xfun 0.52 2025-04-02 [2] CRAN (R 4.4.2)
## yaml 2.3.10 2024-07-26 [2] CRAN (R 4.4.2)
##
## [1] /tmp/RtmpnkJ2cn/temp_libpath5cf253694ba6
## [2] /data/home/kylem/R/x86_64-pc-linux-gnu-library/4.4
## [3] /opt/R/4.4.2/lib/R/library
## * ── Packages attached to the search path.
##
## ──────────────────────────────────────────────────────────────────────────────