Diagnostic Displays
Kyle Baron
2025-10-11
Source:vignettes/diagnostic-displays.Rmd
diagnostic-displays.RmdOverview
This feature set provides standardized displays of diagnostics, including
- ETA versus covariates
- NPDE or CWRES versus covariates
- NPDE or CWRES diagnostics as
- single, comprehensive panel
- just scatter plots
- just histogram and QQ plots
In addition to these standard displays, the user can get an object back containing the component plots for the standardized displays that you can arrange yourself.
library(pmplots)
data <- pmplots_data_obs()
id <- pmplots_data_id()
cont <- c("WT//Weight (kg)", "AGE//Age (years)")
cat <- c("CPc//Child-Pugh", "STUDYc//Study")
covs <- c(cont,cat)
etas <- c("ETA1//ETA-CL", "ETA2//ETA-V", "ETA3//ETA-KA")ETA versus covariates
The basic / default behavior is to get a list of arranged plots, one
for each ETA
p <- eta_covariate(id, x = covs, y = etas)
names(p) ## [1] "ETA1" "ETA2" "ETA3"
p$ETA1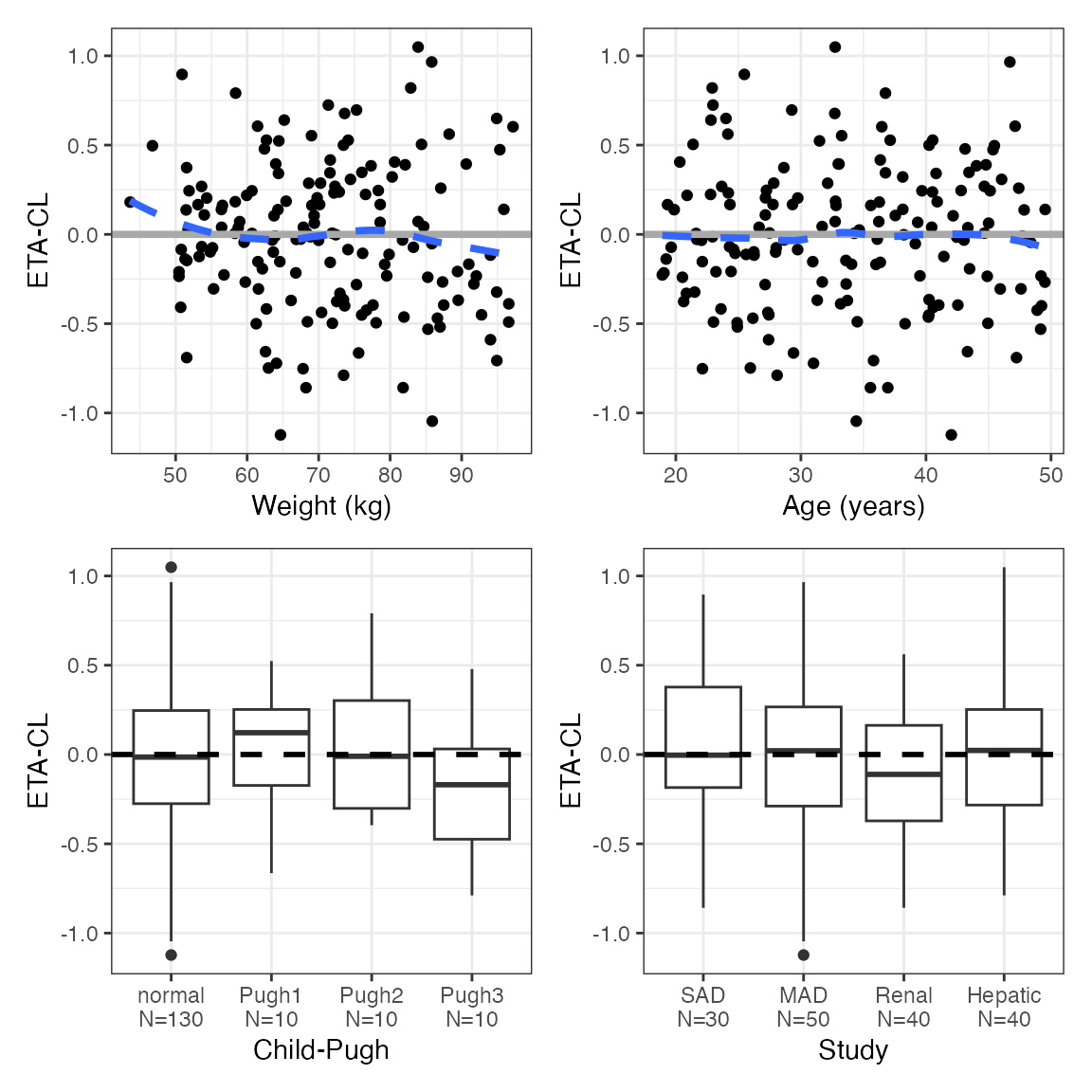
We can label the panels; thinking about making this the default
p <- eta_covariate(id, x = covs, y = etas, tag_levels = "A")
p$ETA1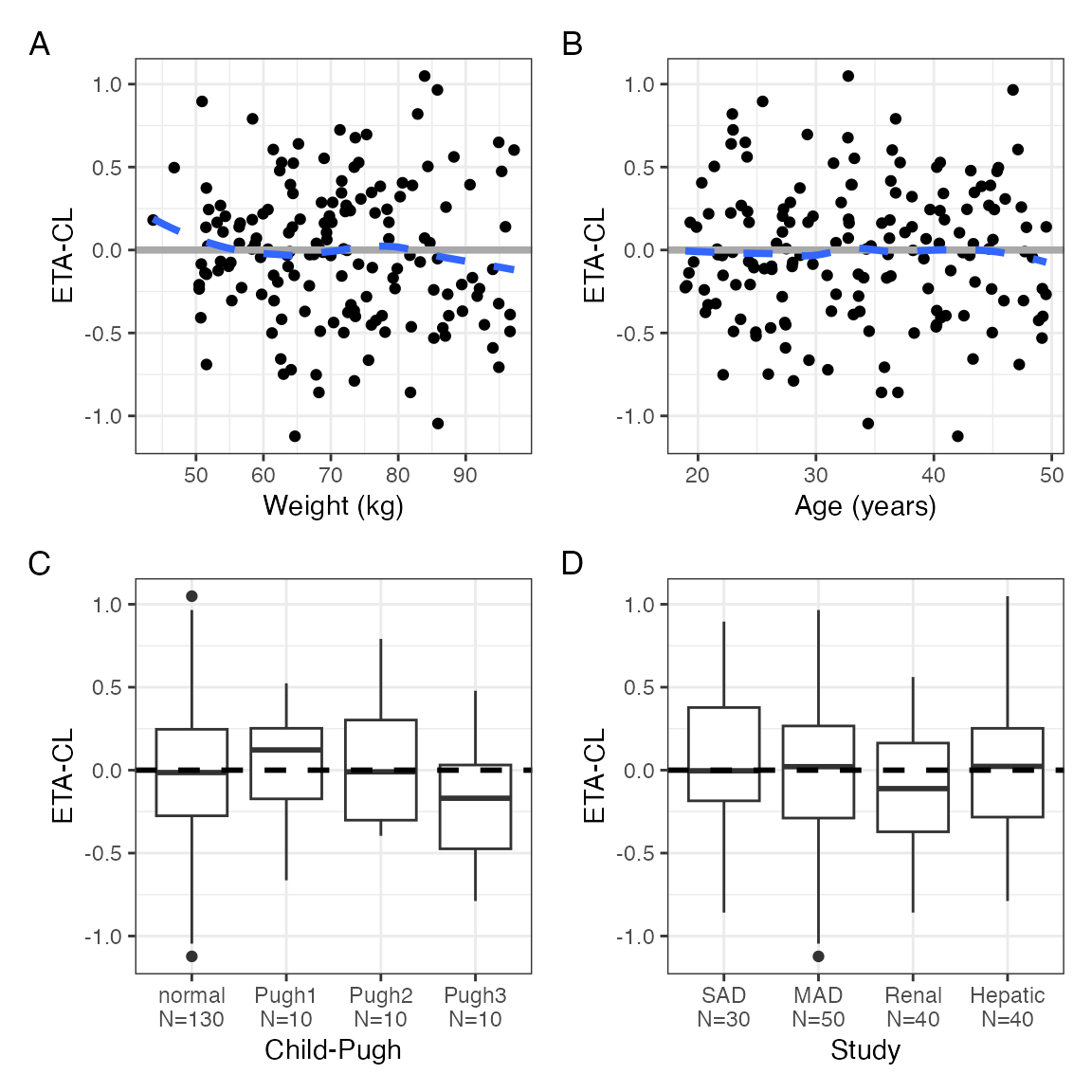
We can arrange this by column instead
p <- eta_covariate(id, x = covs, y = etas, tag_levels = "A", byrow = FALSE)
p$ETA1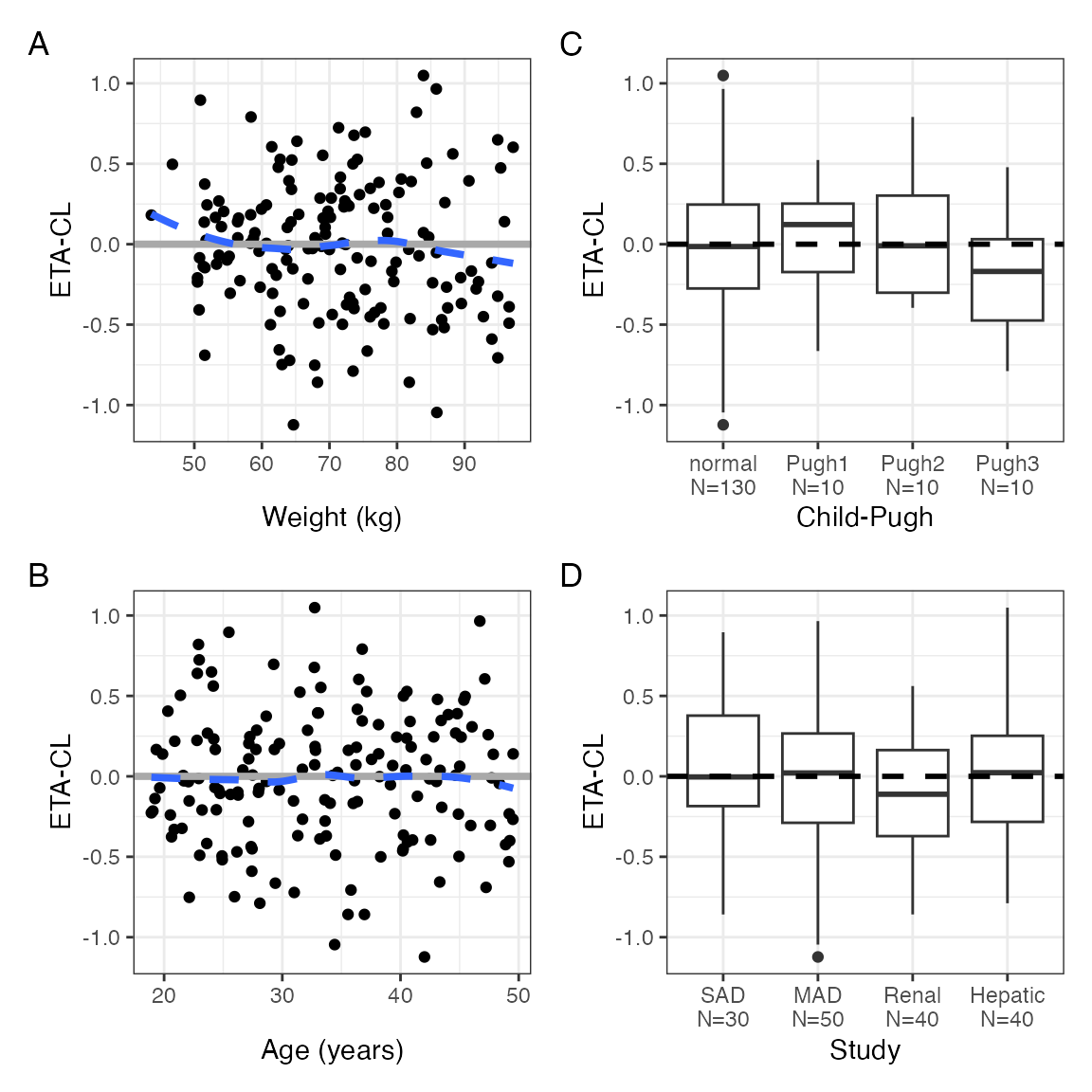
Or we can group by the covariate rather than the ETA
using the transpose argument
p <- eta_covariate(id, x = covs, y = etas, tag_levels = "A", transpose = TRUE)
names(p)## [1] "WT" "AGE" "CPc" "STUDYc"Now, we have all the ETAs for each covariate on the same
page
p$AGE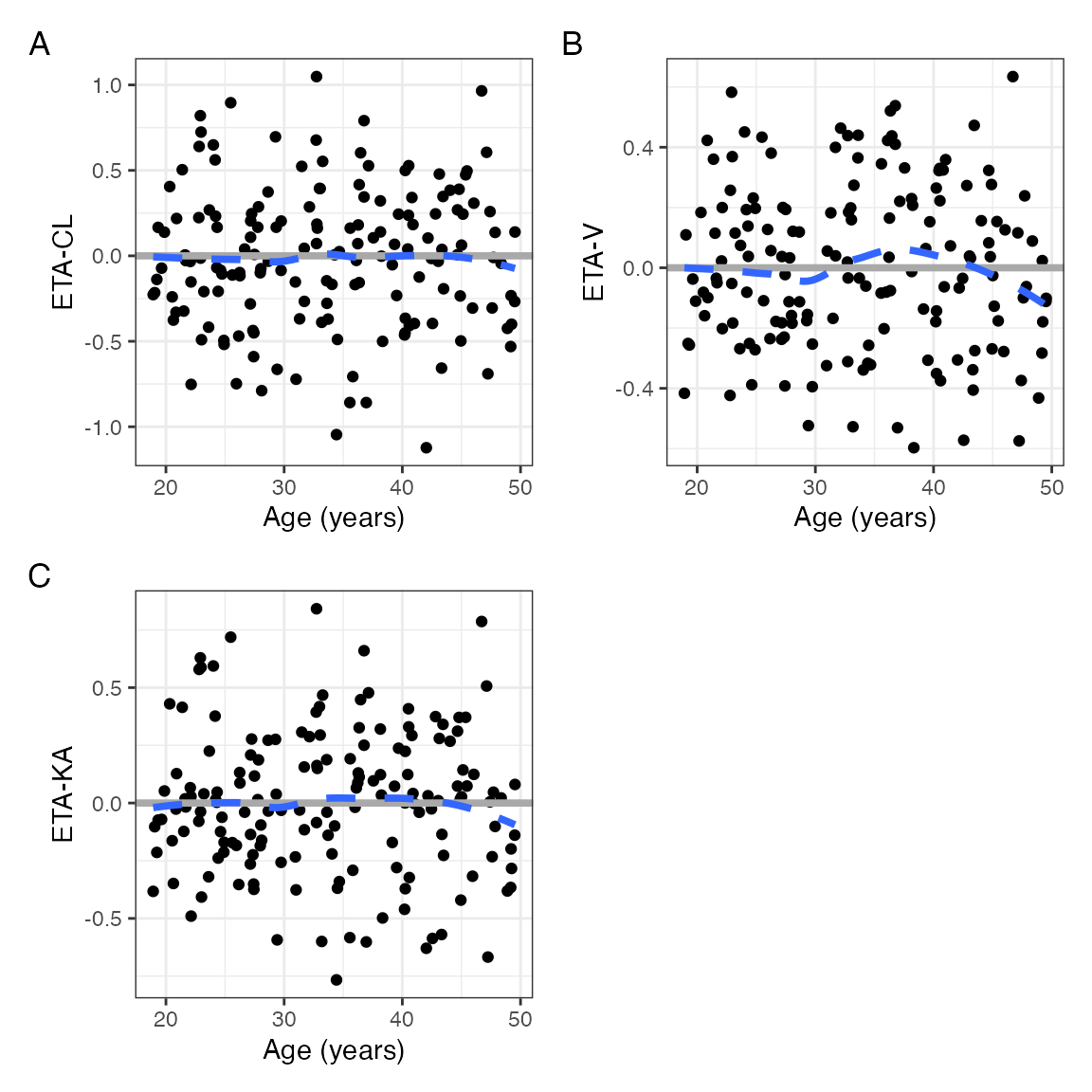
and
p$CPc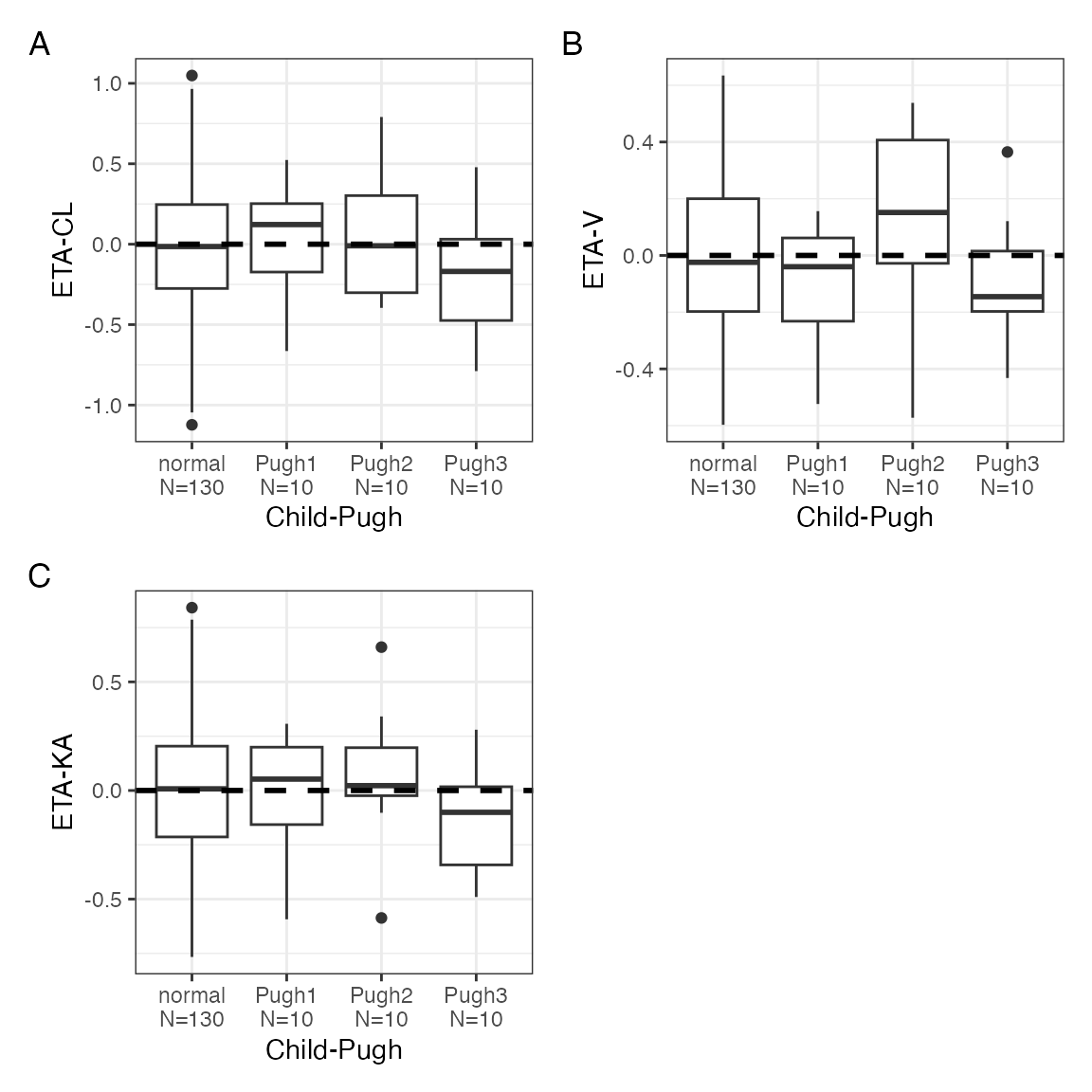
We can make a custom arrangement using the patchwork
arrangement operators
p <- eta_covariate_list(id, x = covs, y = etas)
with(p$ETA1, (WT / (CPc + STUDYc) / AGE), tag_levels = "A")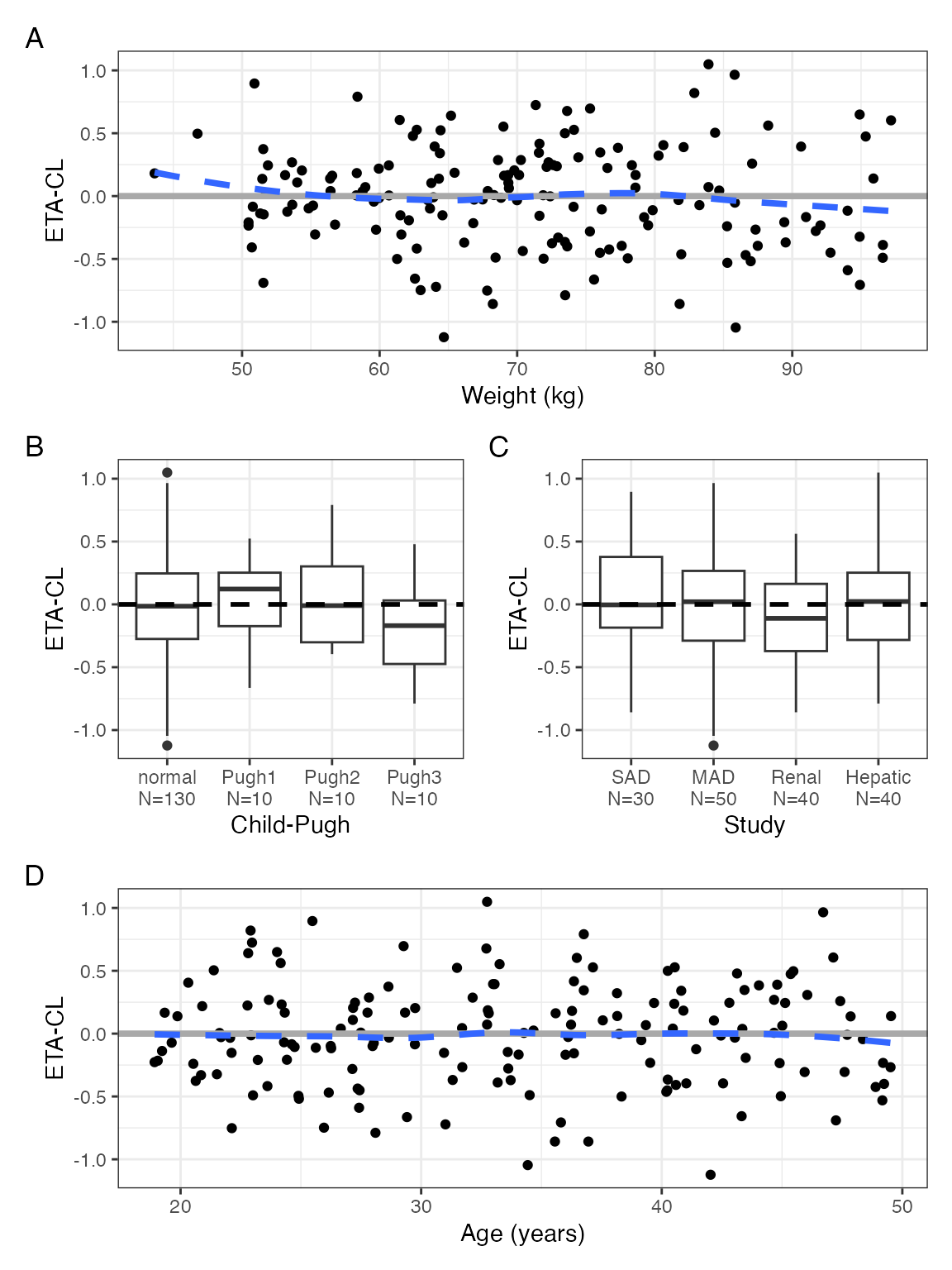
Standard NPDE diagnostics
You can get all the NPDE diagnostics in a single
graphic. This might be too much for a report, but could be handy for
your model checkout script
npde_panel(data, tag_levels = "A")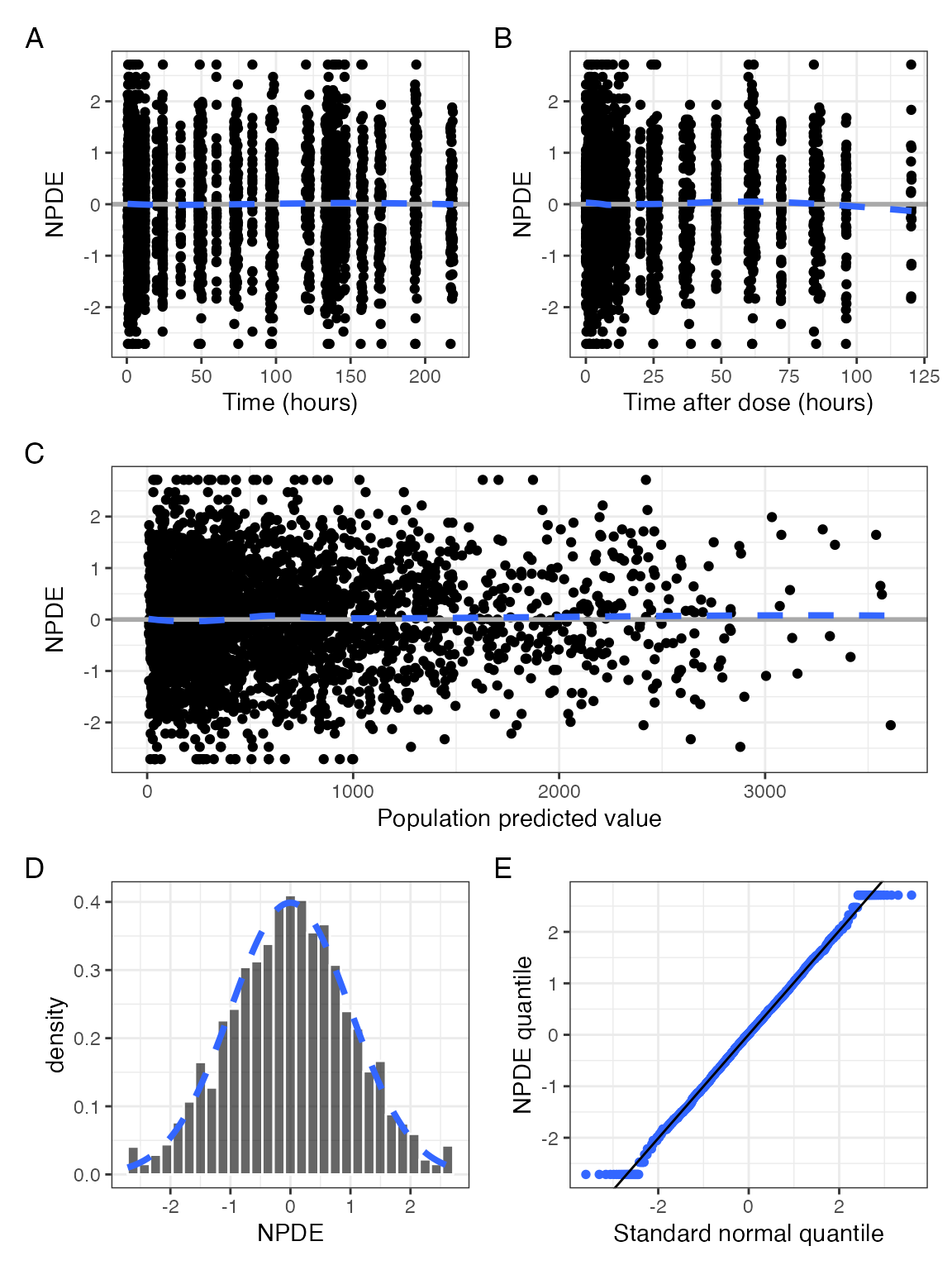
or just the histogram and qq plot
npde_hist_q(data, tag_levels = "A")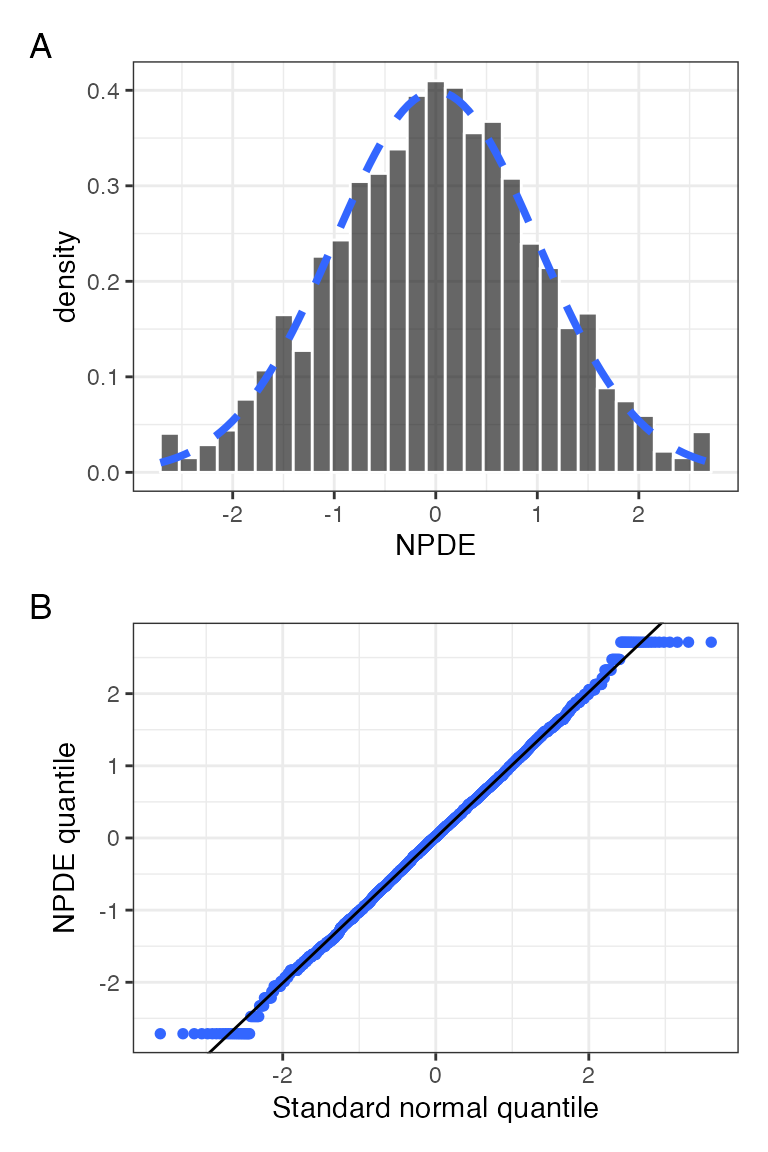
or just the scatter plots in long format
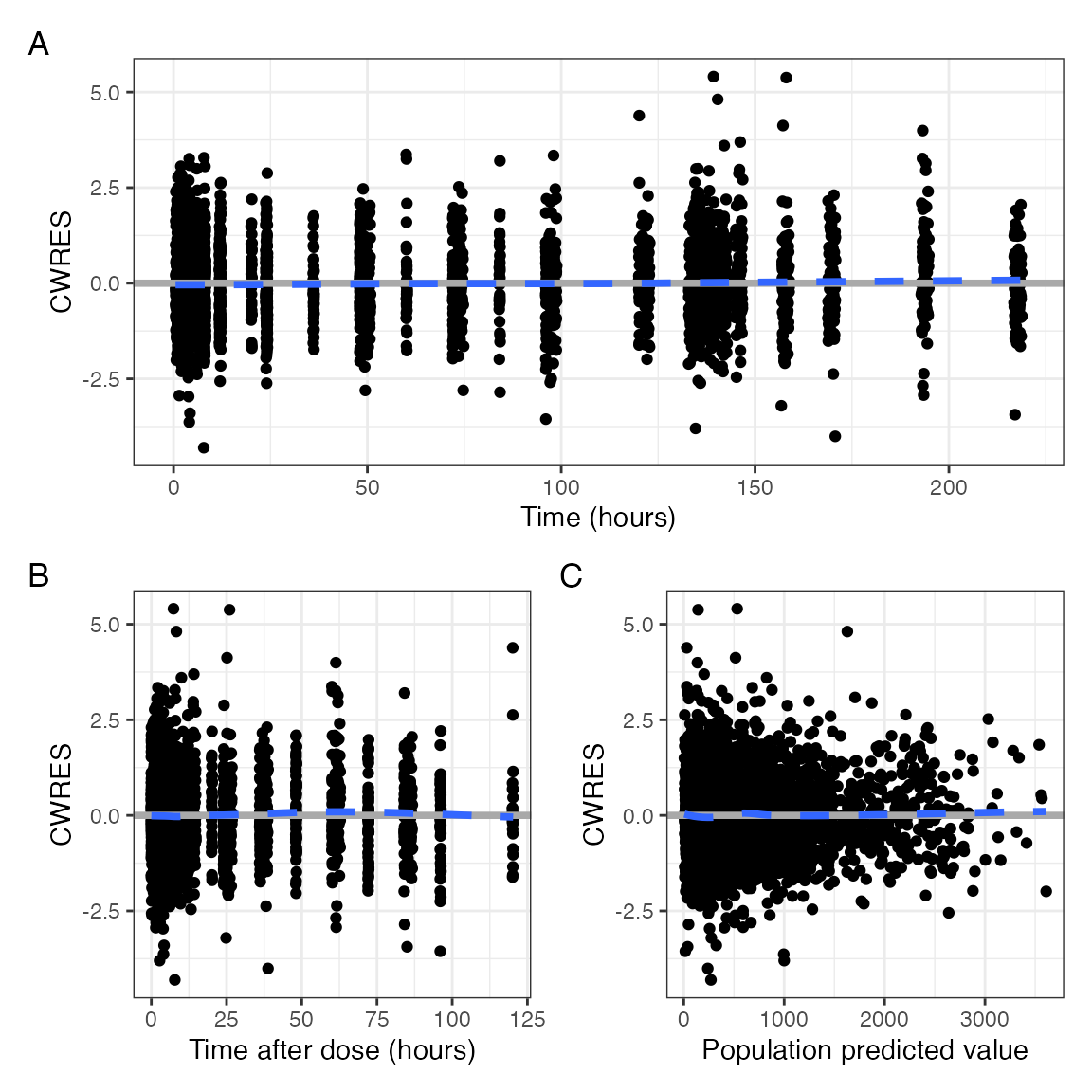
npde_scatter(data, tag_levels = "A")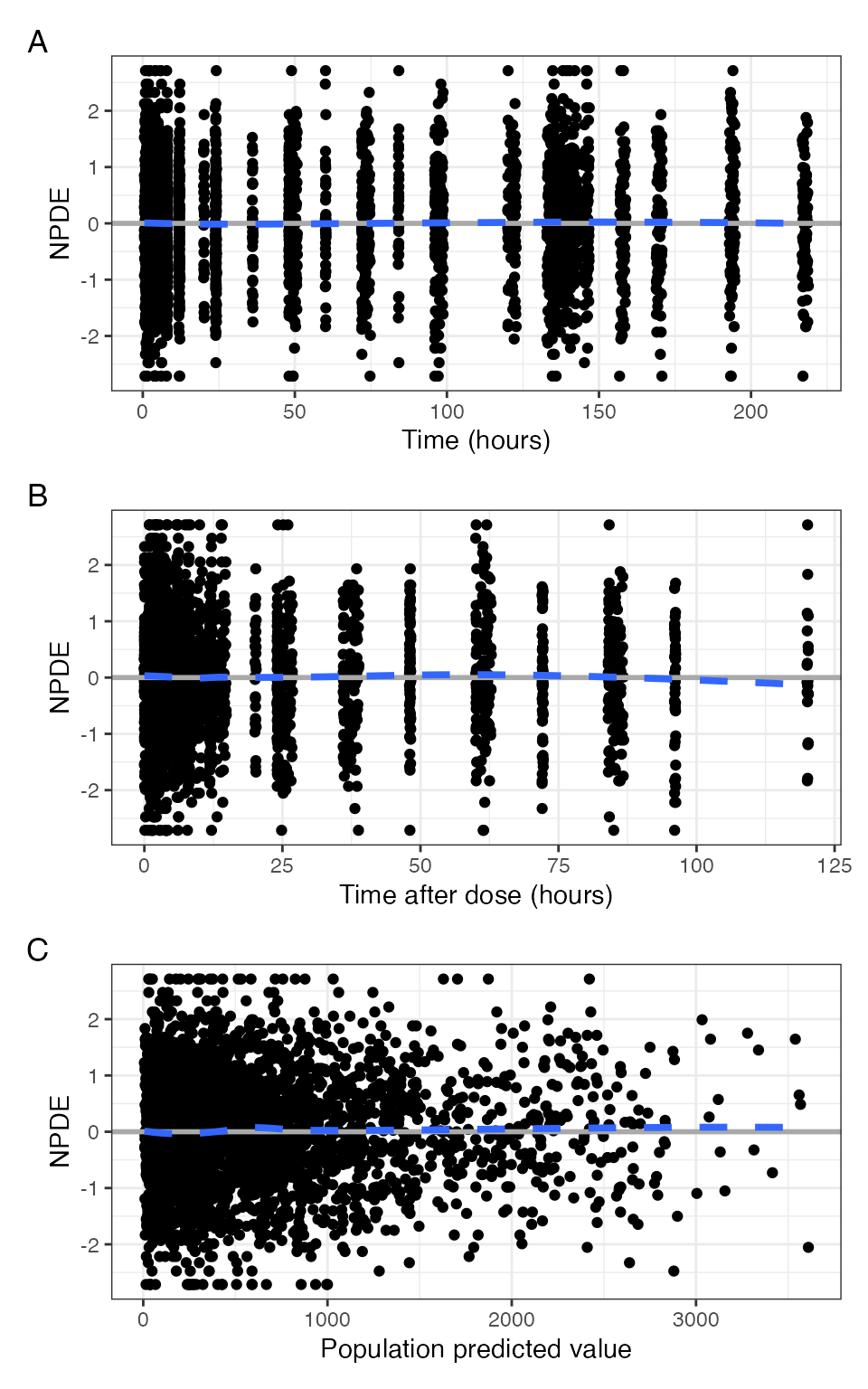
or compact format
npde_scatter(data, tag_levels = "A", compact = TRUE)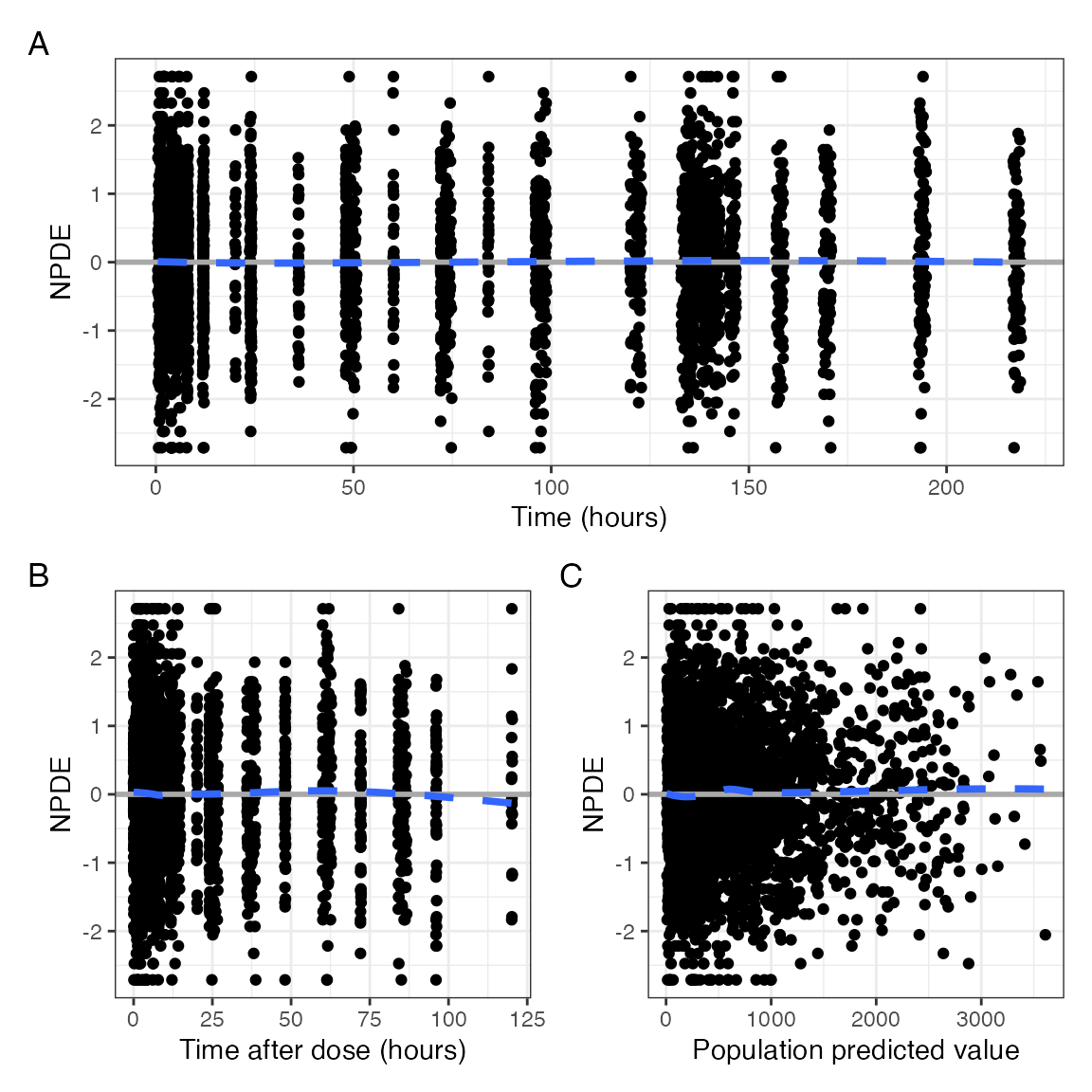
You can also customize the layout to be whatever you want
plots <- npde_panel_list(data)
with(plots, time / (hist + q + tad), tag_levels = "A") 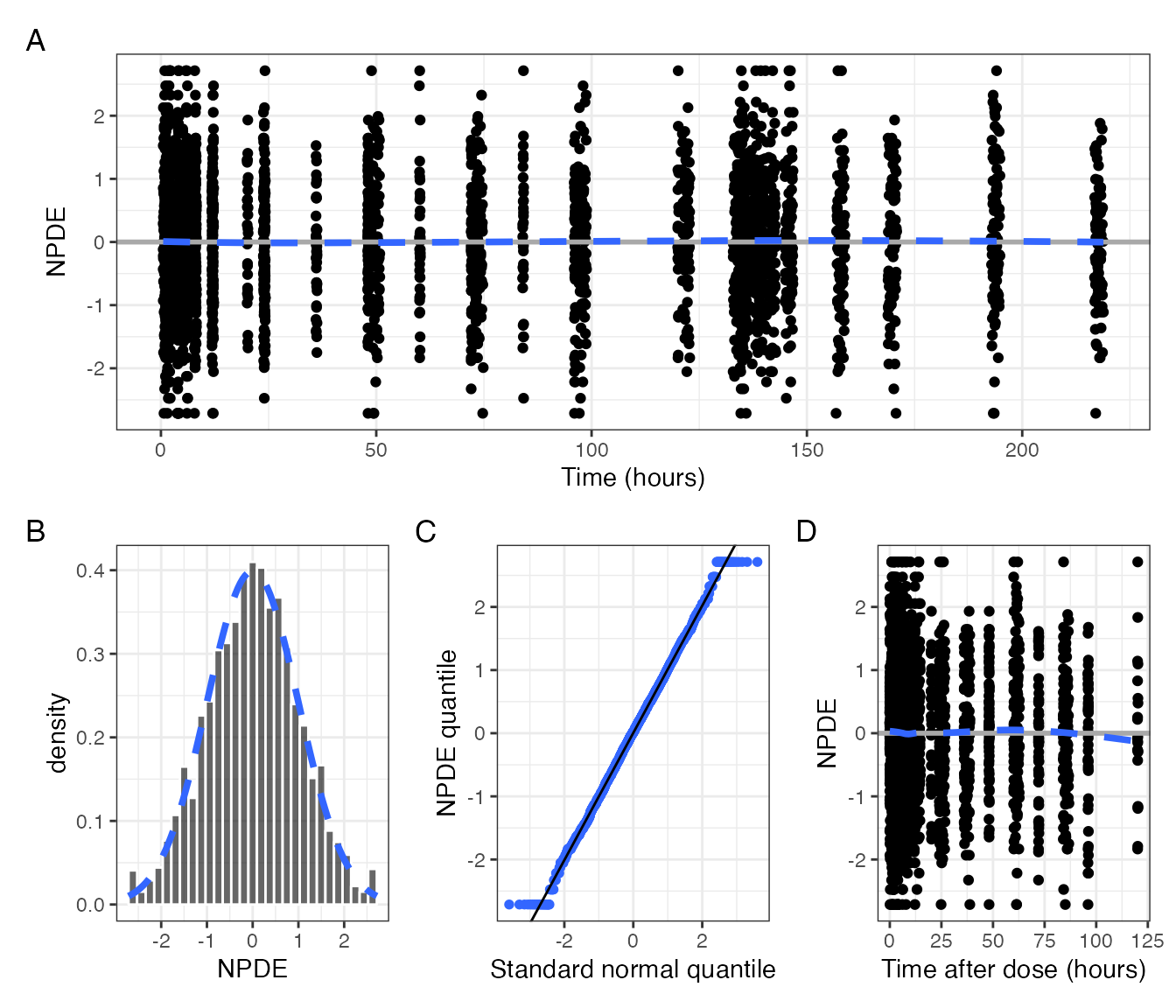
There is some customization for the plot axes and titles, including
- x-axis tick intervals for
timeandtad - x-axis units for
timeandtad - the name for
pred
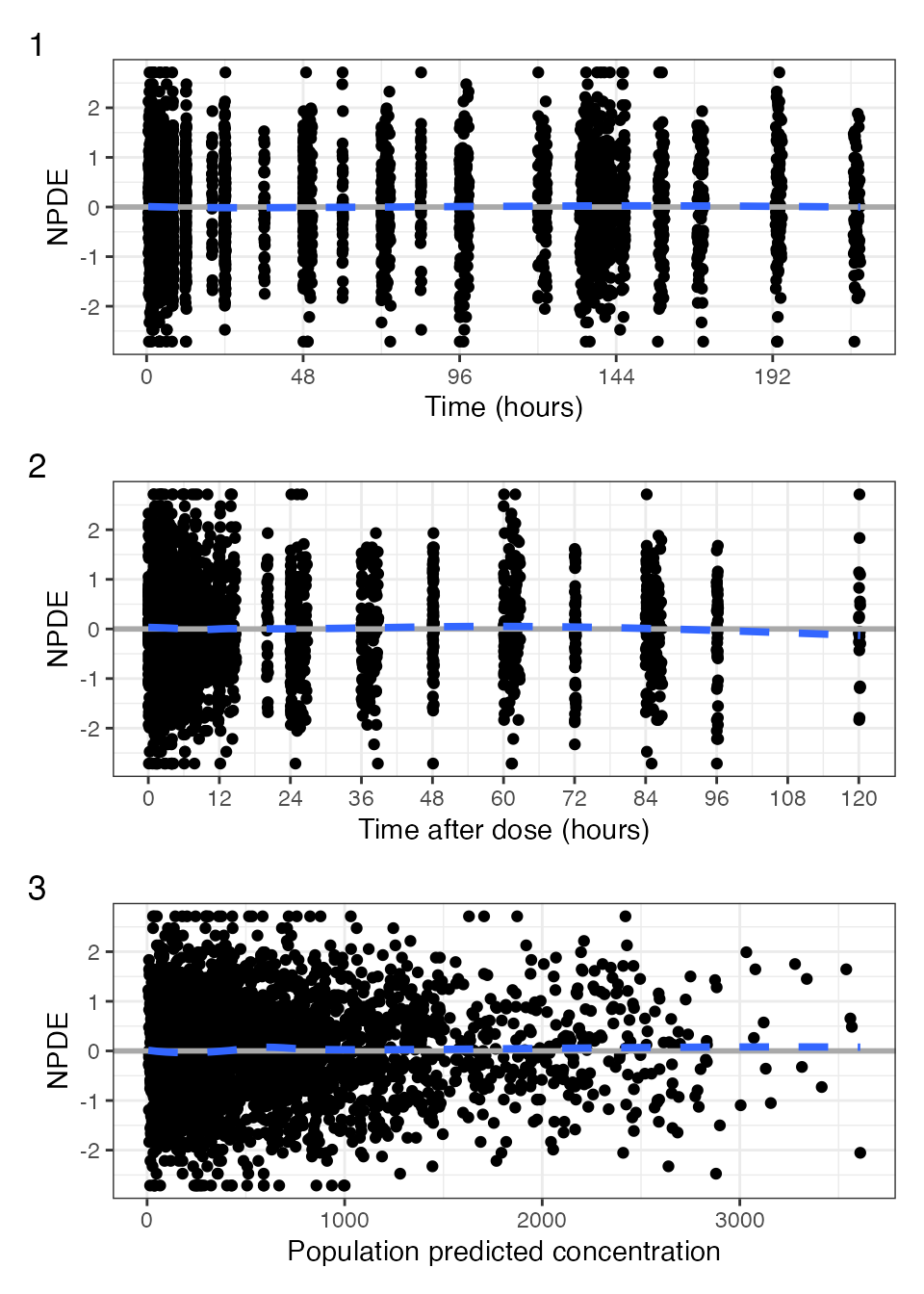
npde_scatter(
data,
xby_tad = 12,
xby_time = 48,
xname = "concentration",
tag_levels = 1
)
NPDE versus covariates
This works like eta_covariate()
npde_covariate(data, covs, tag_levels = "a", byrow = FALSE)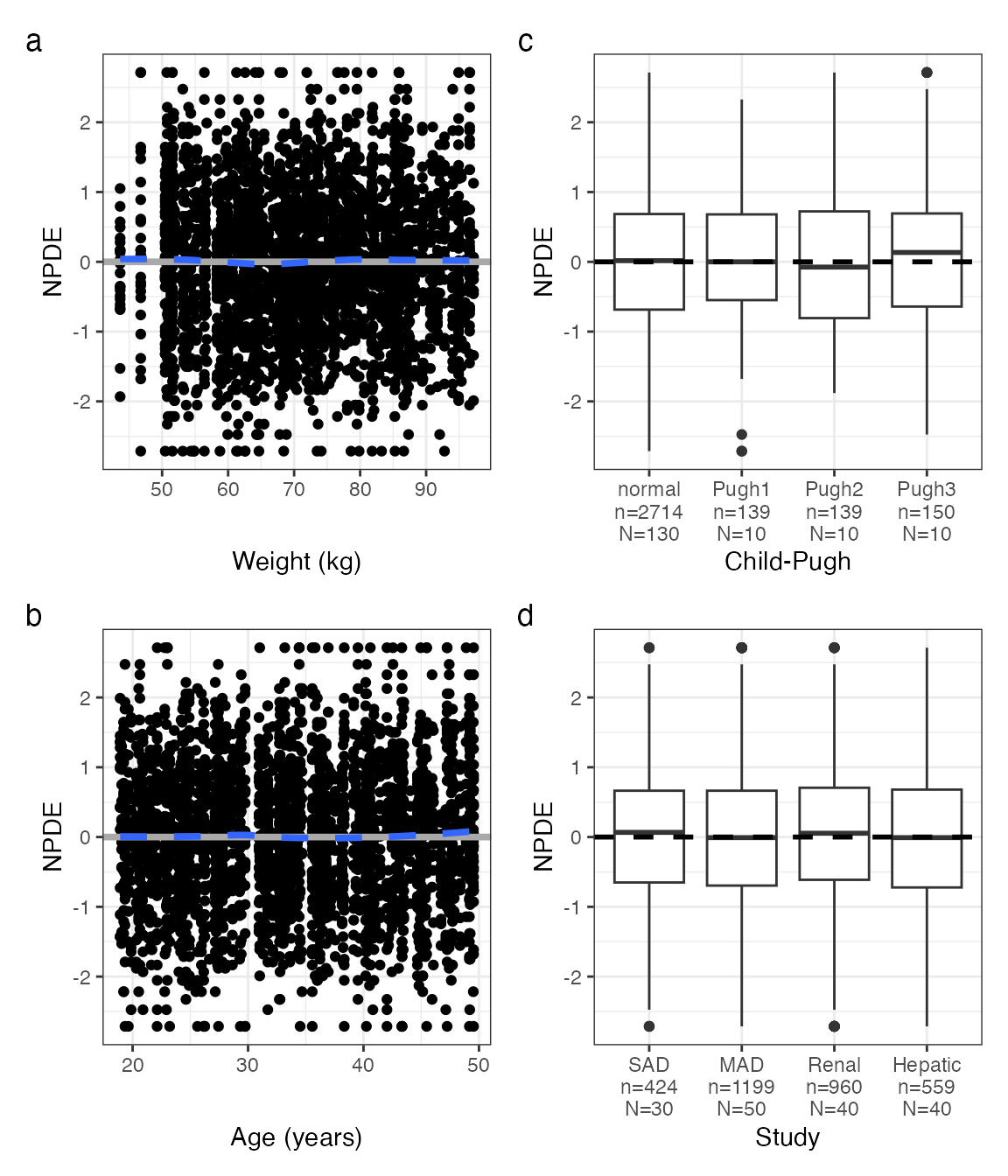
plots <- npde_covariate_list(data, covs)
names(plots)## [1] "WT" "AGE" "CPc" "STUDYc"
with(plots, (AGE + WT) / CPc / STUDYc, tag_levels = "i")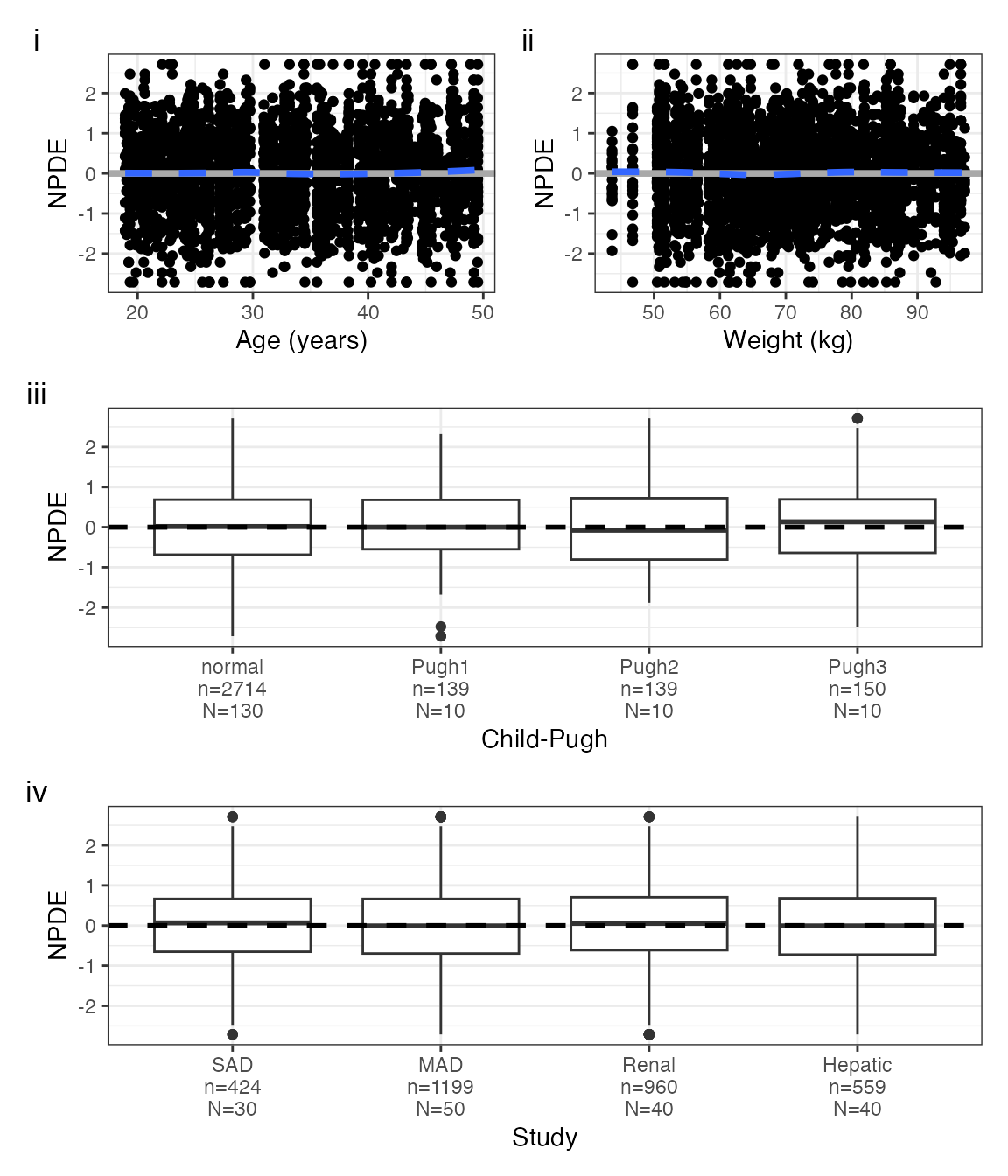
Standard CWRES diagnostics
The behavior / feature set looks just like the NPDE
plots for both standard diagnostics and covariates.
cwres_scatter(data, tag_levels = "A", compact = TRUE)