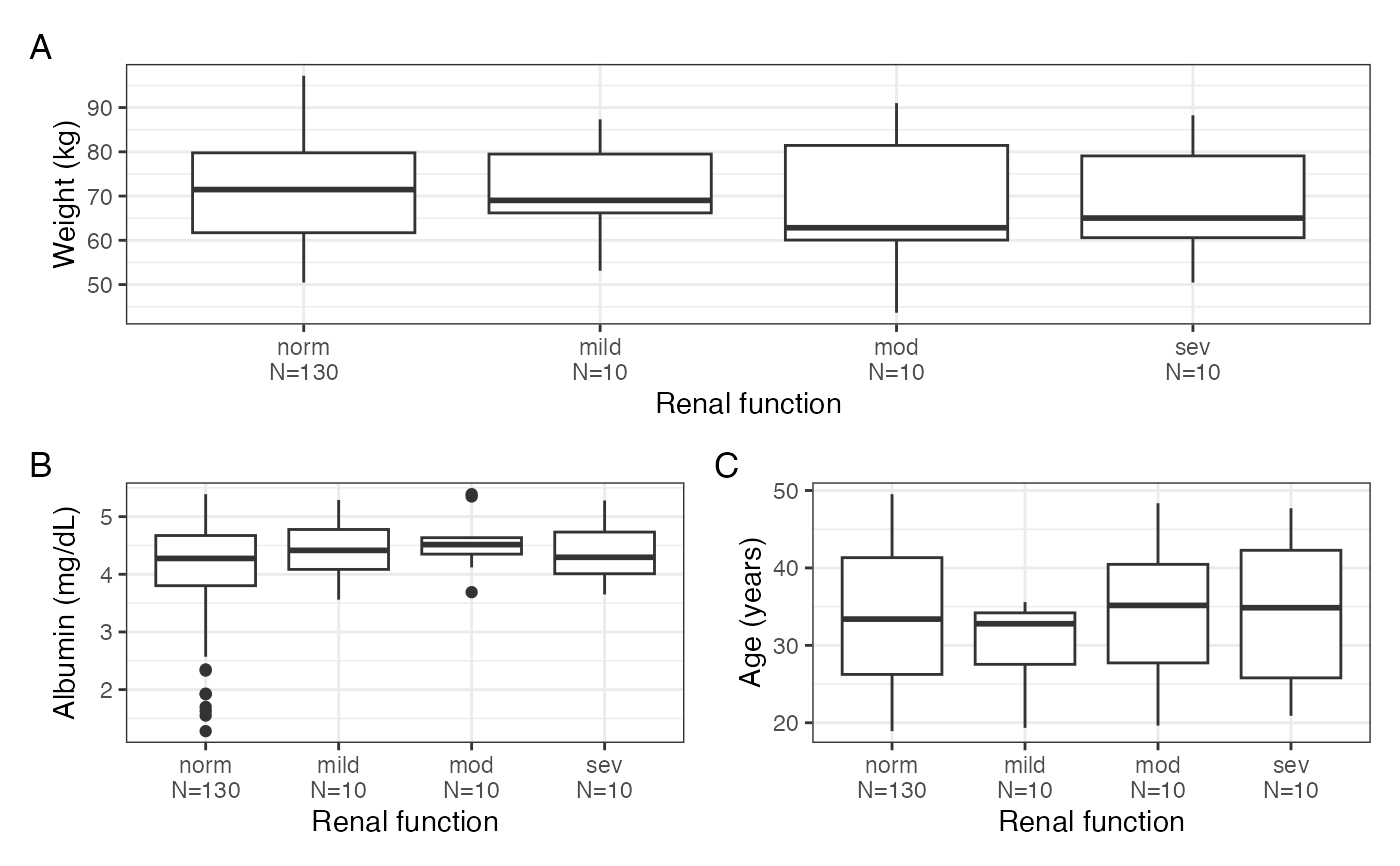
Get a single graphic of continuous covariate boxplots split by
levels of different categorical covariates (cont_cat_panel()).
Alternatively, get the component plots to be arranged by the user
(cont_cat_panel_list())
cont_cat_panel(
df,
x,
y,
ncol = 2,
tag_levels = NULL,
byrow = FALSE,
transpose = FALSE,
...
)
cont_cat_panel_list(df, x, y, transpose = FALSE, ...)Arguments
- df
a data frame to plot.
- x
character
col//titlefor the categorical covariates to plot on x-axis; seecol_label().- y
character
col//titlefor the continuous covariates to plot on y-axis; seecol_label().- ncol
passed to
pm_grid().- tag_levels
passed to
patchwork::plot_annotation().- byrow
passed to
pm_grid().- transpose
logical; if
TRUE, output will be transposed to group plots by the categorical covariates rather than the continuous covariates.- ...
additional arguments passed to
cont_cat().
Value
cont_cat_panel() returns a list of plots arranged in graphics as a
patchwork object using pm_grid(). cont_cat_panel_list() returns the
same plots, but unarranged as a named list of lists.
When transpose is FALSE (default), plots in a single graphic are grouped
by the continuous covariates (passed as y), and the names of the list
reflect those names (e.g., WT). When transpose is TRUE, the graphics
are grouped by the categorical covariates (passed as x) and the names of
the list reflect those names (e.g. RF). See Examples.
Details
Pass ncol = NULL or another non-numeric value to bypass arranging plots
coming from cont_cat_panel().
See also
Examples
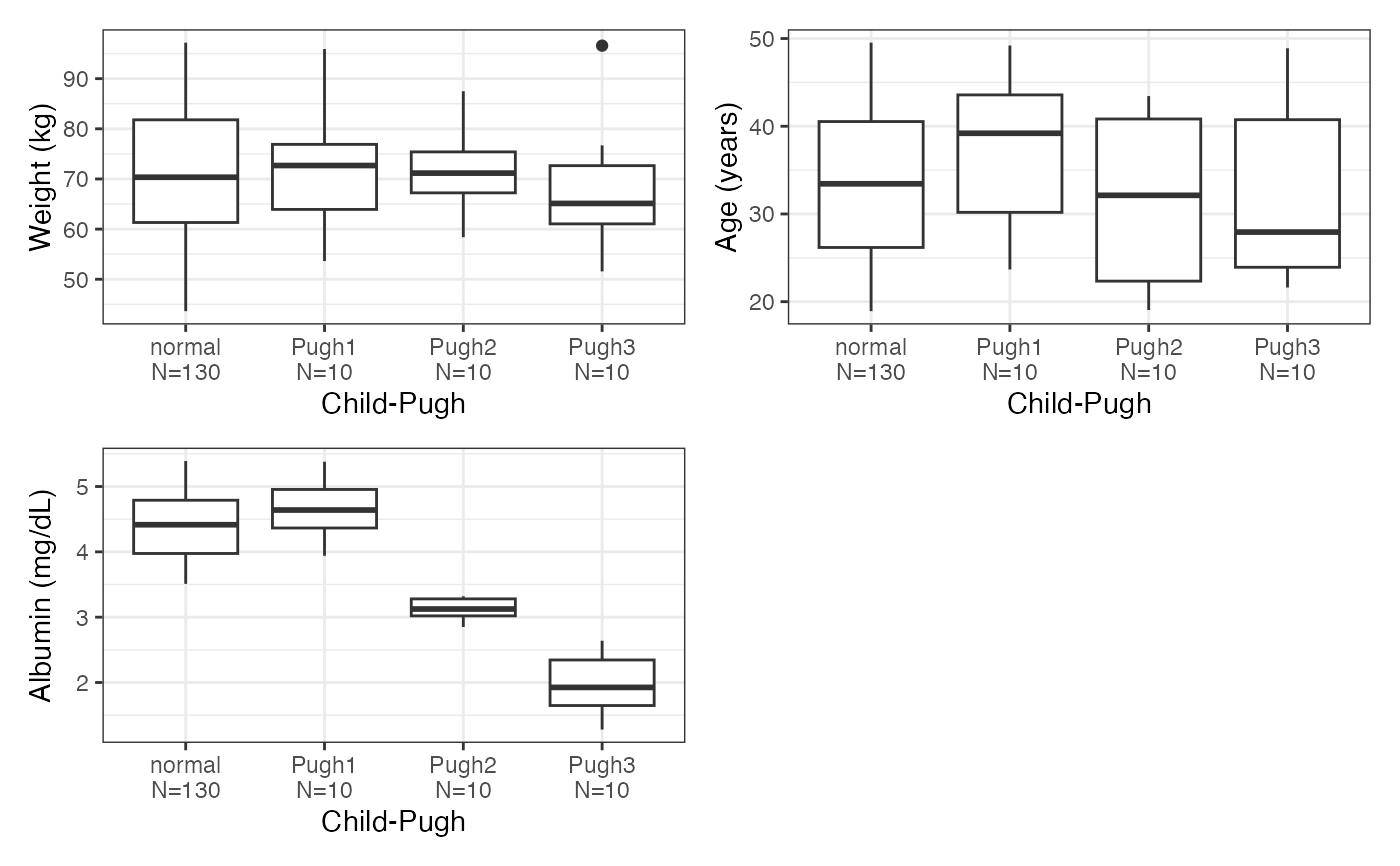
data <- pmplots_data_id()
cont <- c("WT//Weight (kg)", "ALB//Albumin (mg/dL)", "AGE//Age (years)")
cats <- c("RF//Renal function", "CPc//Child-Pugh")
cont_cat_panel(data, x = cats, y = cont, tag_levels = "A")
#> $WT
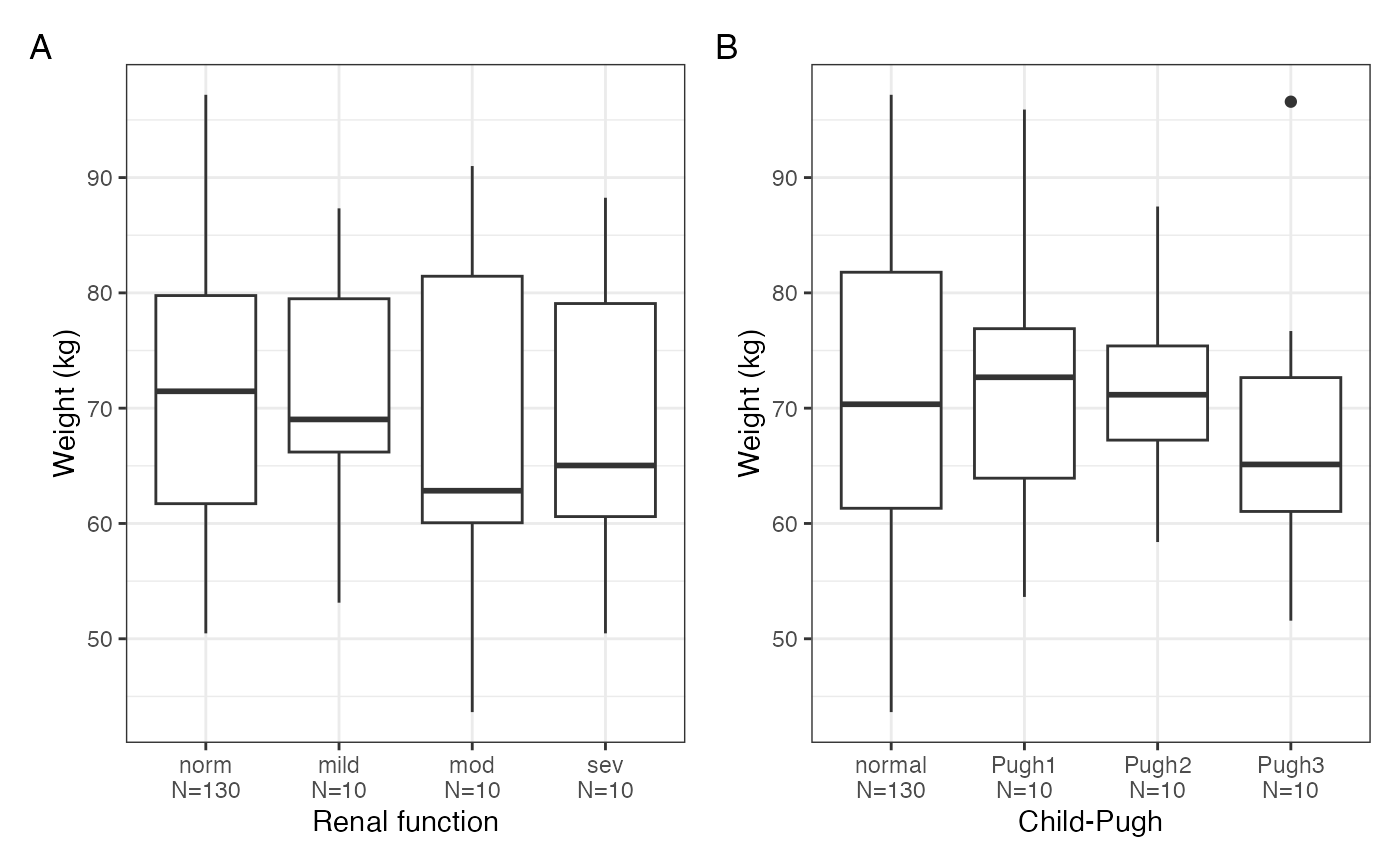 #>
#> $ALB
#>
#> $ALB
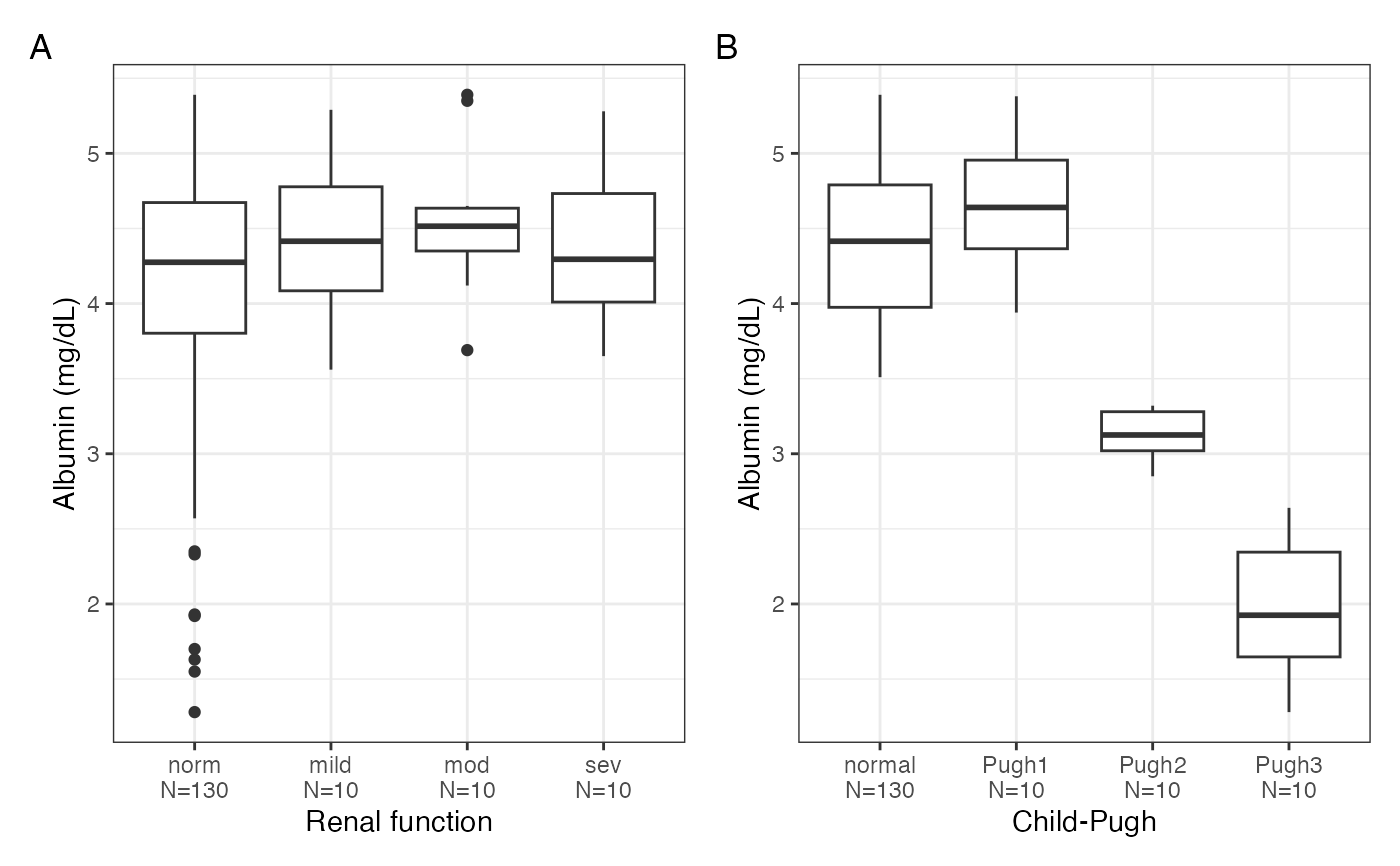 #>
#> $AGE
#>
#> $AGE
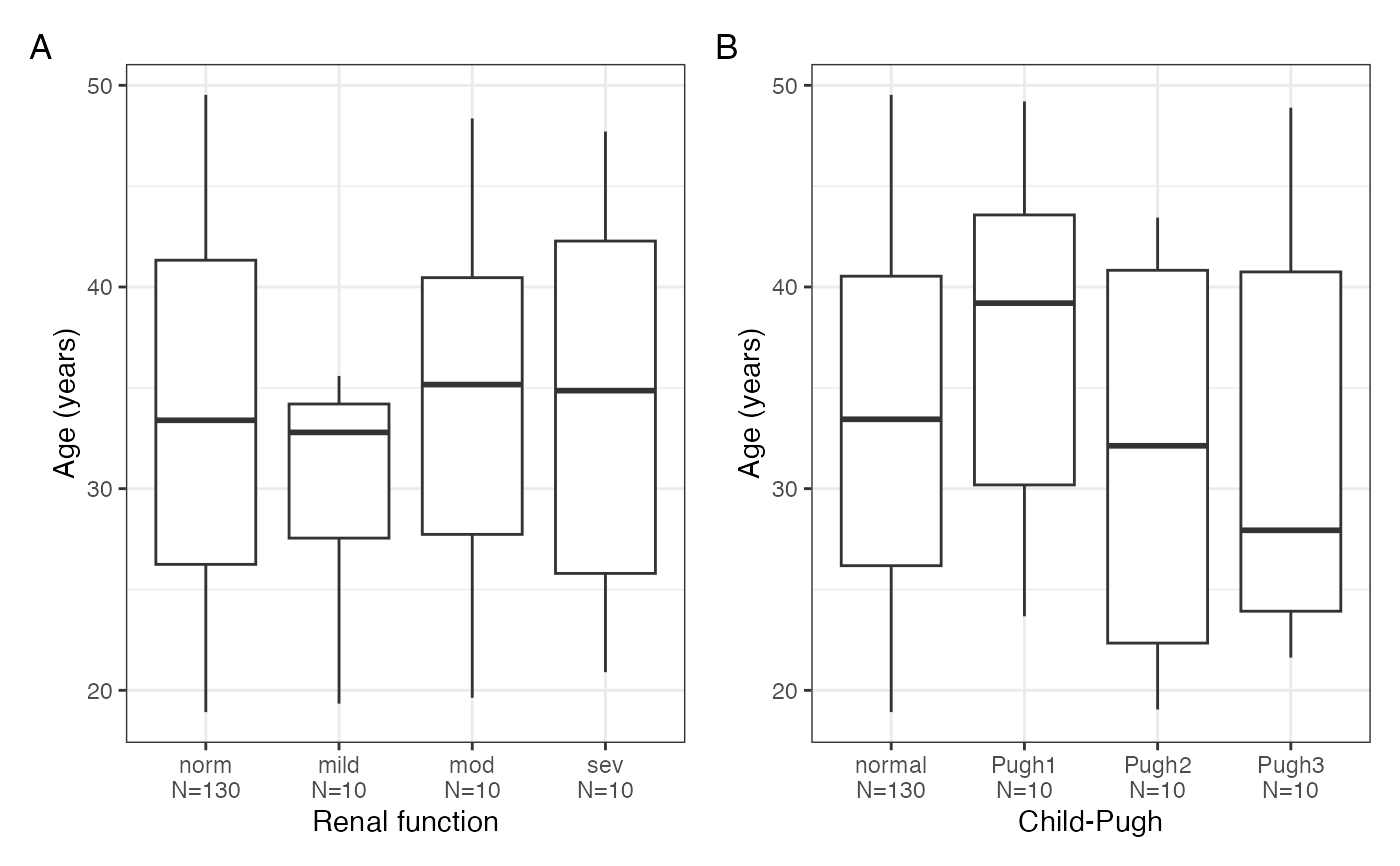 #>
cont_cat_panel(data, cats, cont)
#> $WT
#>
cont_cat_panel(data, cats, cont)
#> $WT
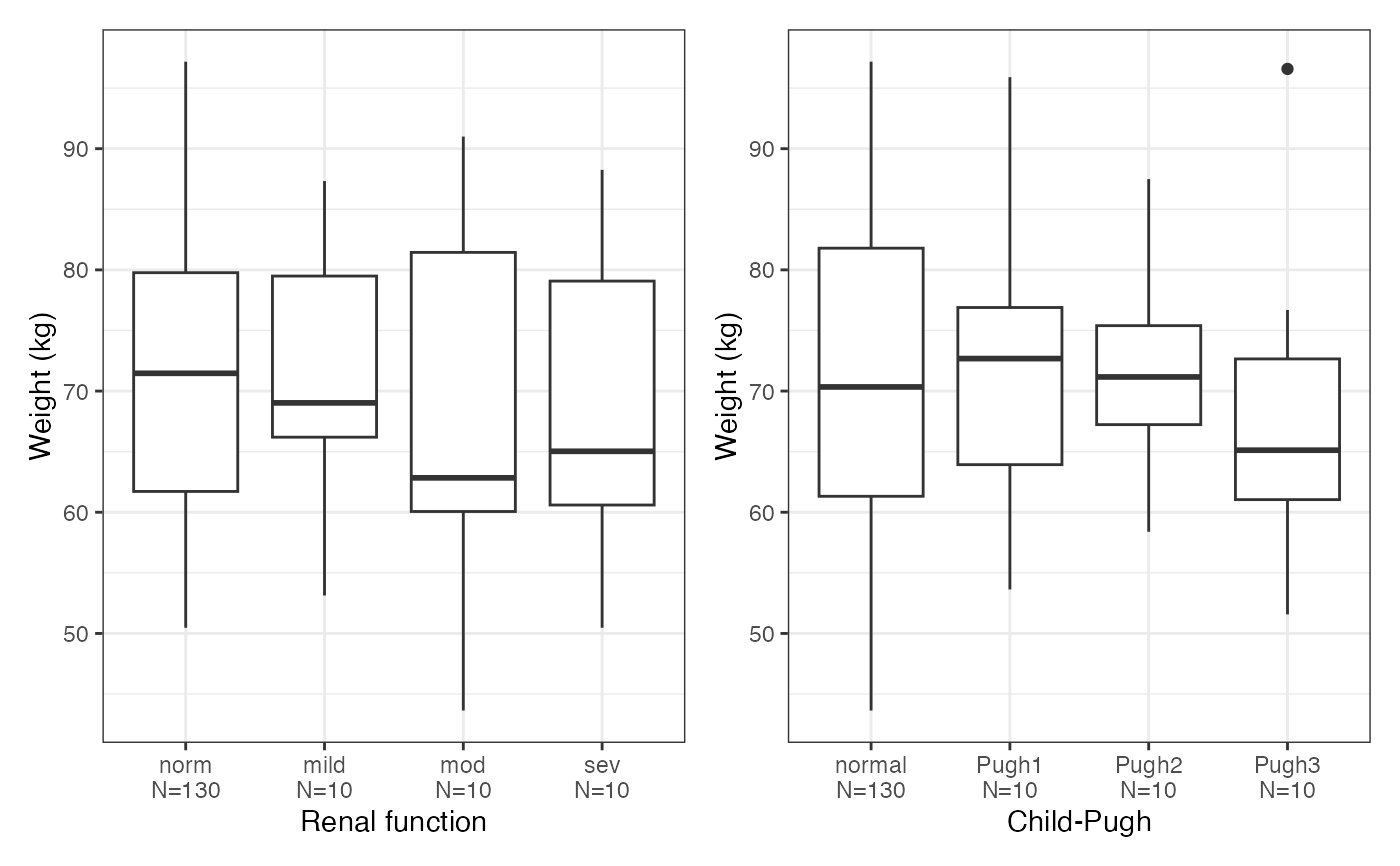 #>
#> $ALB
#>
#> $ALB
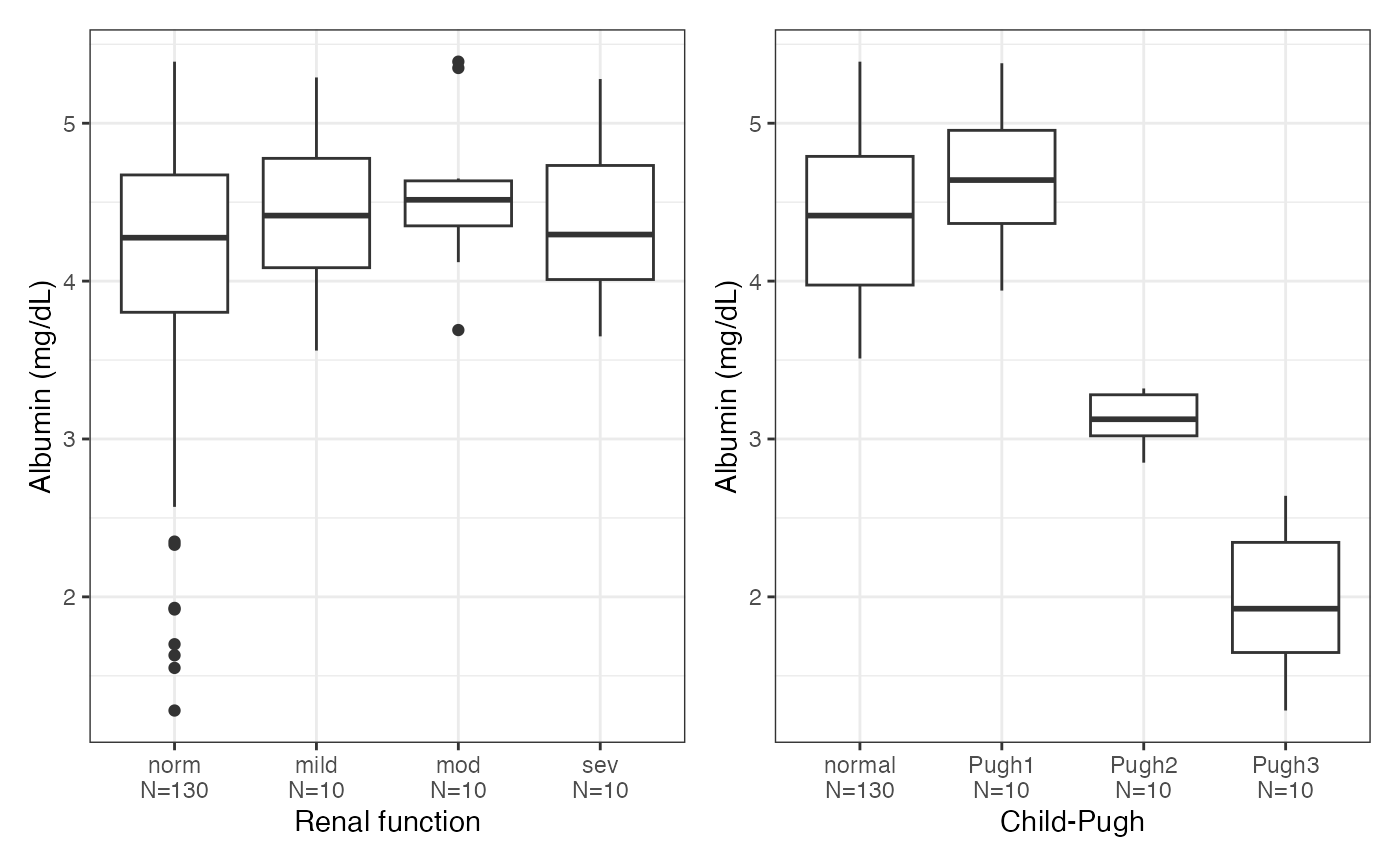 #>
#> $AGE
#>
#> $AGE
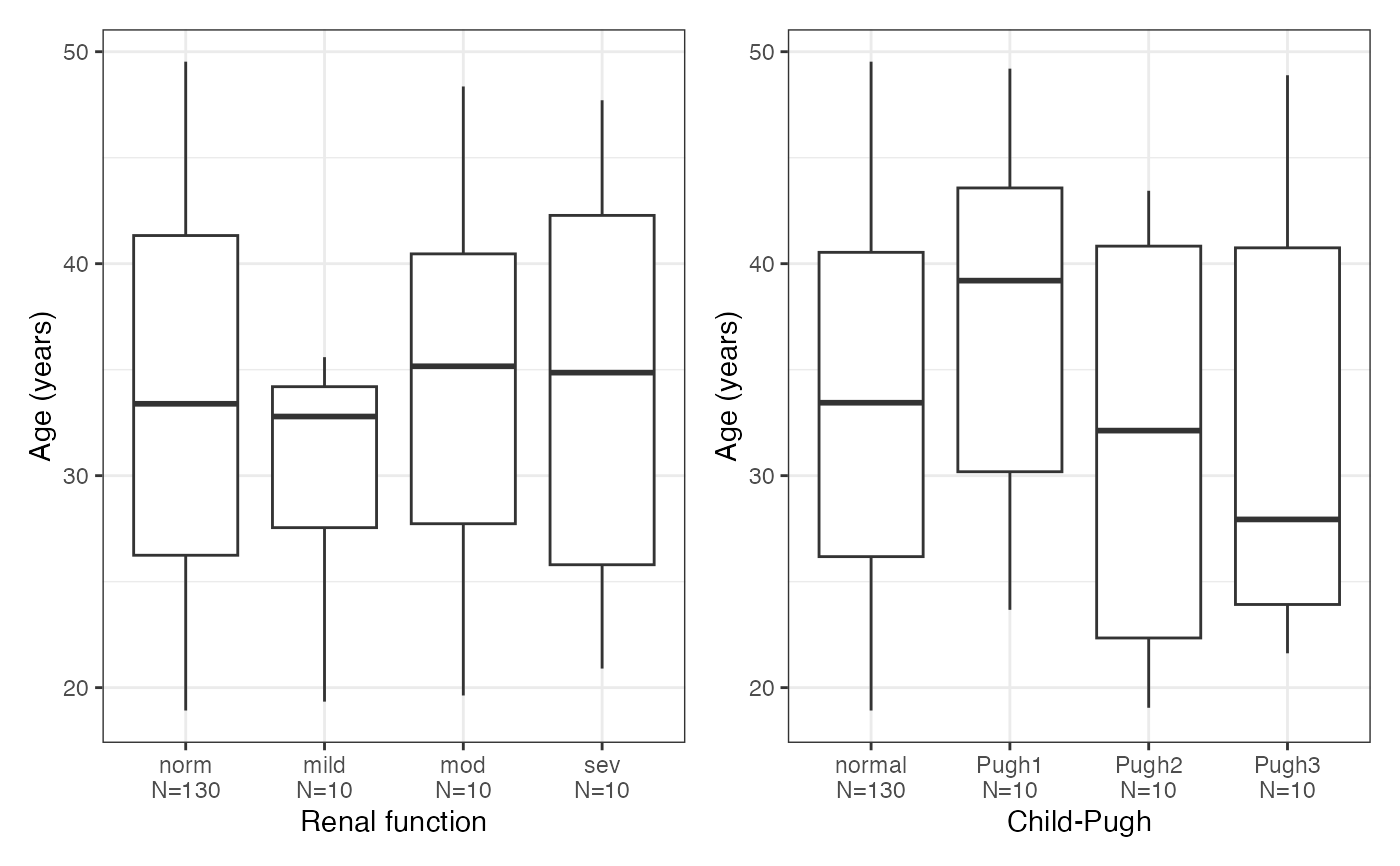 #>
cont_cat_panel(data, cats, cont, transpose = TRUE)
#> $RF
#>
cont_cat_panel(data, cats, cont, transpose = TRUE)
#> $RF
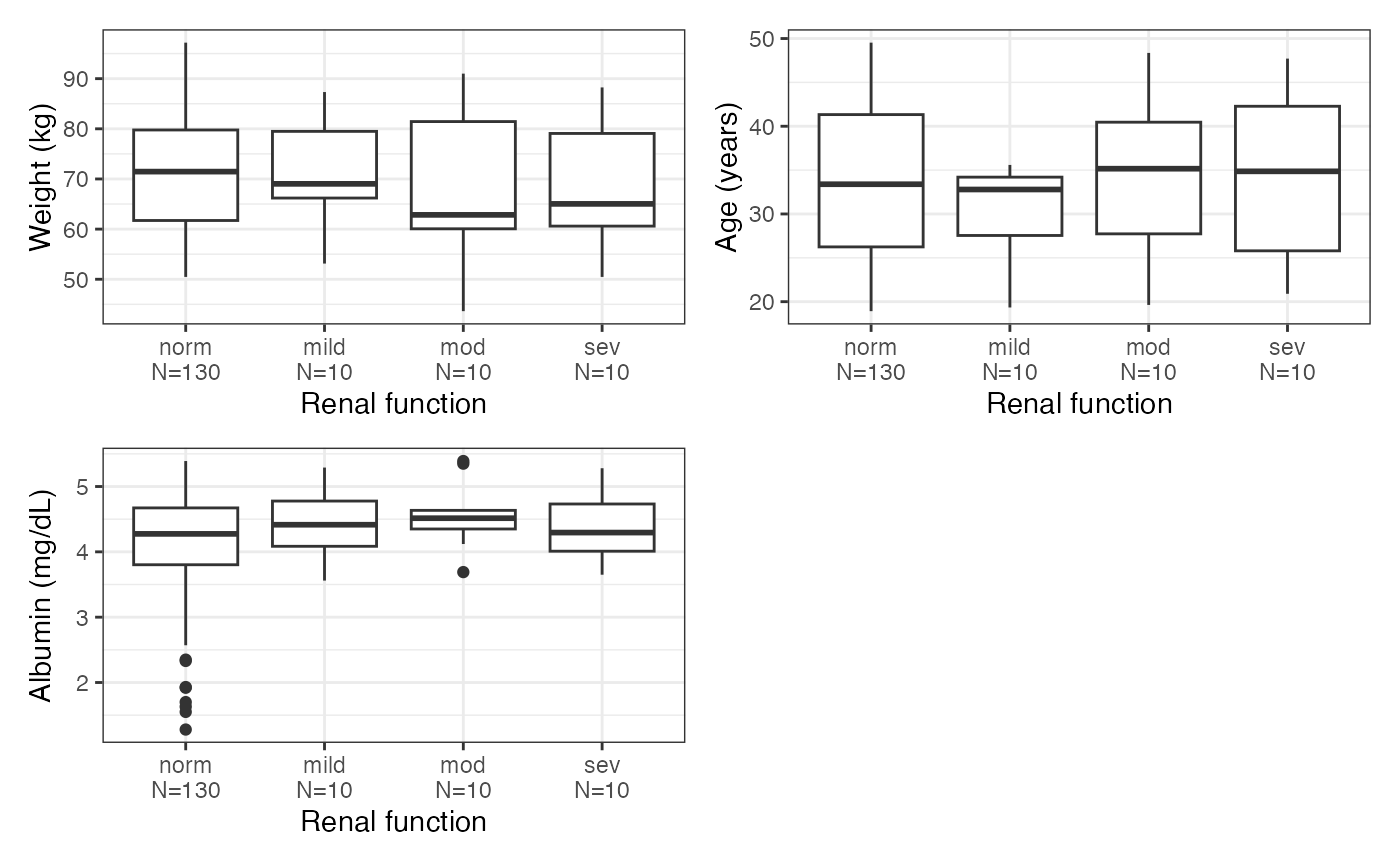 #>
#> $CPc
#>
#> $CPc
 #>
l <- cont_cat_panel_list(data, cats, cont, transpose = TRUE)
names(l)
#> [1] "RF" "CPc"
with(l$RF, WT/(ALB + AGE), tag_levels = "A")
#>
l <- cont_cat_panel_list(data, cats, cont, transpose = TRUE)
names(l)
#> [1] "RF" "CPc"
with(l$RF, WT/(ALB + AGE), tag_levels = "A")