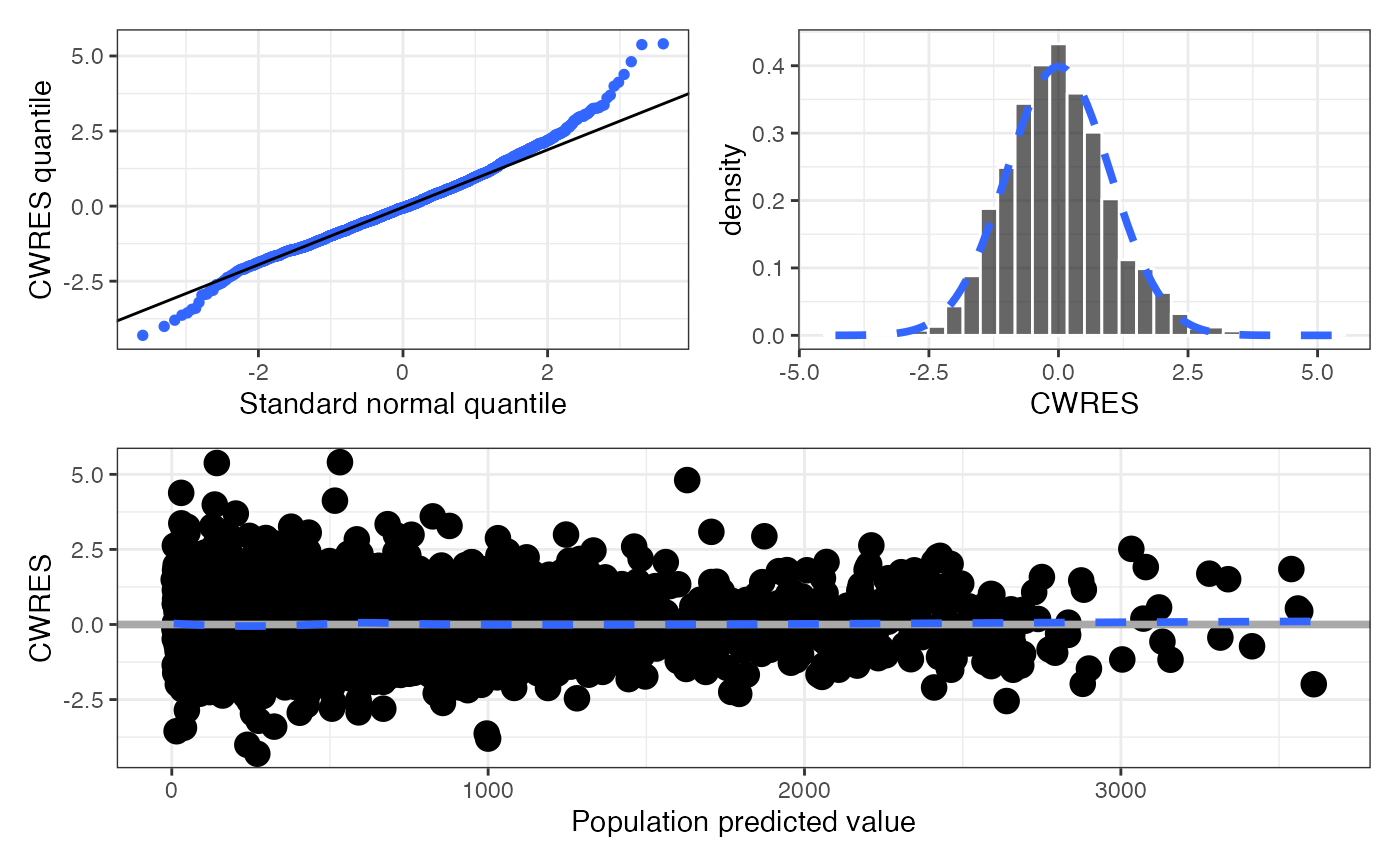
Get a single graphic of basic CWRES diagnostics (cwres_panel()) or get
the component plots in a list that can be arranged by the user
(cwres_panel_list()) . See cwres_covariate() for plotting CWRES
versus covariates.
cwres_panel(
df,
xname = "value",
unit_time = "hours",
unit_tad = "hours",
xby_time = NULL,
xby_tad = NULL,
tag_levels = NULL
)
cwres_panel_list(
df,
xname = "value",
unit_time = "hours",
unit_tad = "hours",
xby_time = NULL,
xby_tad = NULL
)Arguments
- df
a data frame to plot.
- xname
passed to
npde_pred().- unit_time
passed to
npde_tad()asxunit.- unit_tad
passed to
npde_time()asxunit.- xby_time
passed to
npde_time()asxby.- xby_tad
passed to
npde_tad()asxby.- tag_levels
passed to
patchwork::plot_annotation().
Value
cwres_panel() returns a single graphic with the following panels:
CWRESversusTIMEviacwres_time()CWRESversusTADviacwres_tad()CWRESversusPREDviacwres_pred()CWREShistogram viacwres_hist()CWRESquantile-quantile plot viacwres_q()
cwres_panel_list() returns a list of the individual plots that are
incorporated into the cwres_panel() output. Each element of the list
is named for the plot in that position: time, tad, pred, hist
q. See Examples for how you can work with that list.
See also
Examples
data <- pmplots_data_obs()
cwres_panel(data, tag_levels = "A")
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> `geom_smooth()` using formula = 'y ~ x'
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
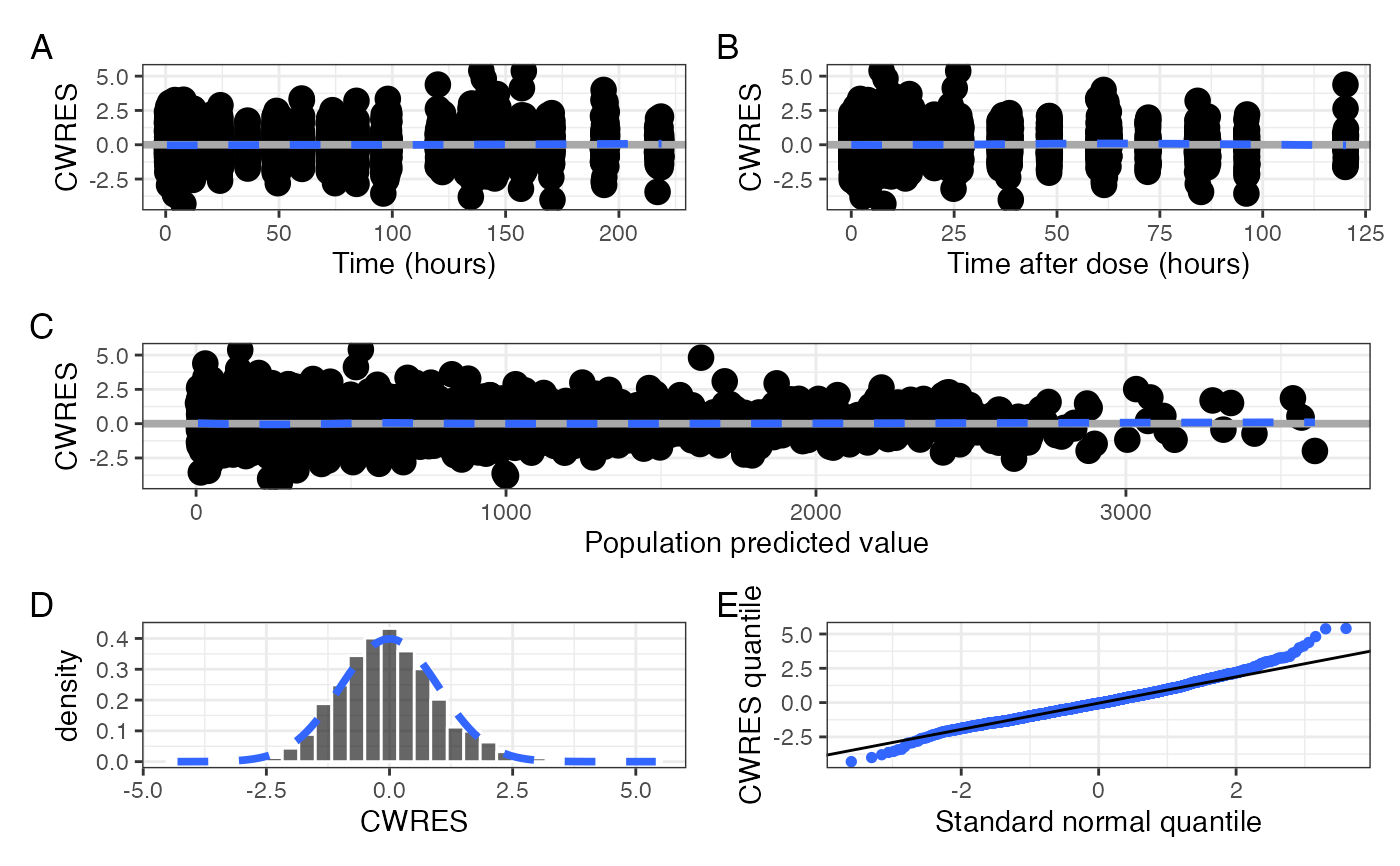 l <- cwres_panel_list(data)
with(l, (q+hist) / pred)
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> `geom_smooth()` using formula = 'y ~ x'
l <- cwres_panel_list(data)
with(l, (q+hist) / pred)
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> `geom_smooth()` using formula = 'y ~ x'